Outcomes of the First AI4Life Open Call
by Beatriz Serrano-Solano & Florian Jug
We are thrilled to announce that we received an impressive number of 72 applications to the first AI4Life Open Call!
The first AI4Life Open Call was launched in mid-February and closed on March 31st, 2023. It is part of the first of a series of three that will be launched over the course of the AI4Life project.
AI4Life involves partners with different areas of expertise, and it covers a range of topics, including marine biology, plant phenotyping, compound screening, and structural biology. Since the goal of AI4Life is to bridge the gap between life science and computational methods, we are delighted to see so much interest from different scientific fields seeking support to tackle scientific analysis problems with Deep Learning methods.
We noticed that the most prominent scientific field that applied was cell biology, but we also received applications from neuroscience, developmental biology, plant ecology, agronomy, and marine biology, as well as aspects of the medical and biomedical fields, such as cardiovascular and oncology.
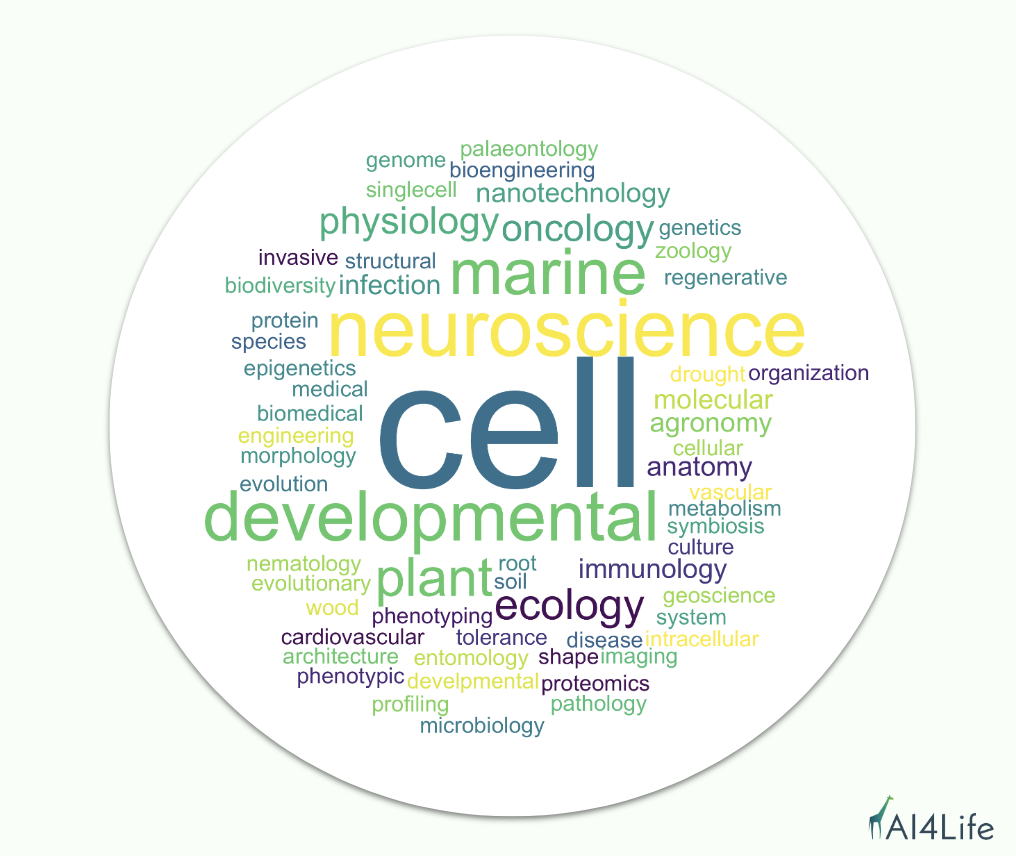
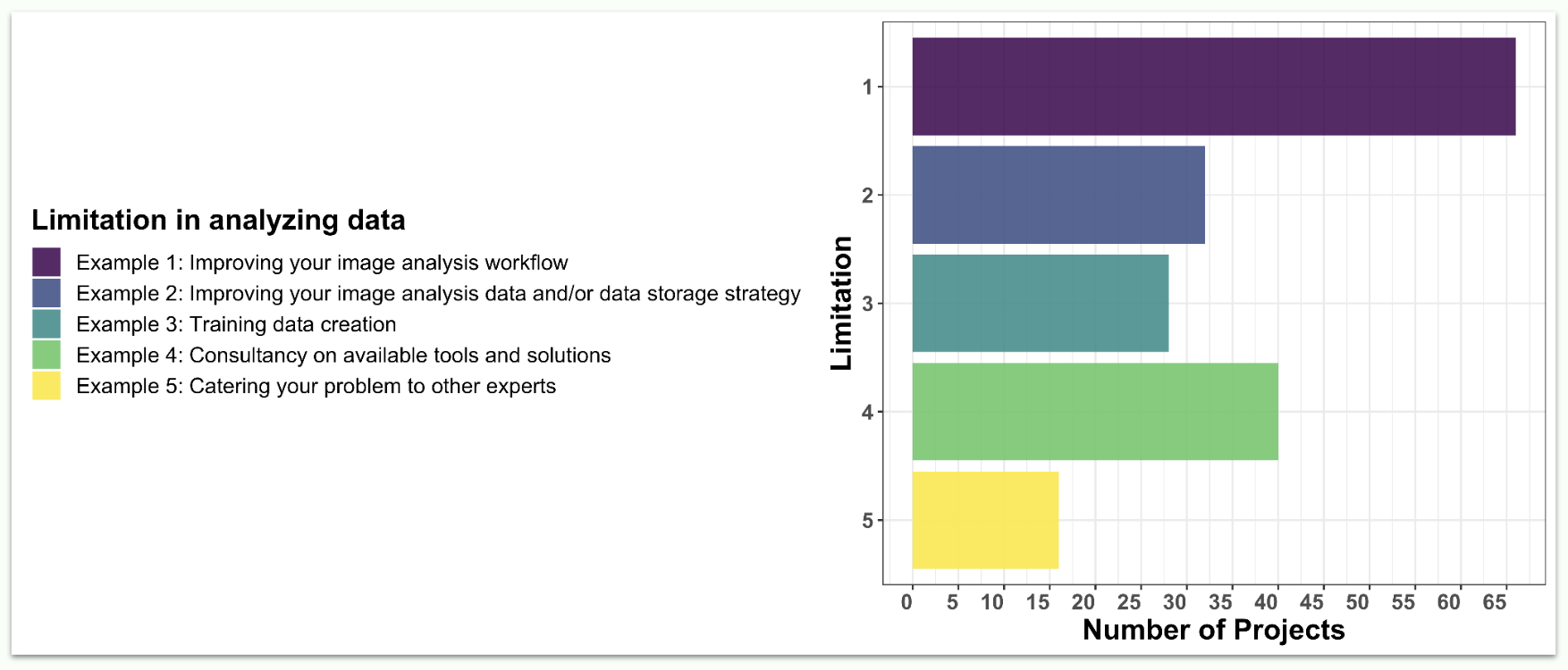
Why did scientists apply? What challenges are they facing?
Applications were classified based on the type of problem that will need to be addressed. We found that improving the applicant’s image analysis workflow was the most common request (67 applications out of 72). This was followed by improving image analysis data and/or data storage strategy, training data creation, and consultancy on available tools and solutions. Of course, we also welcomed more specific challenges that were not falling within any of those categories.
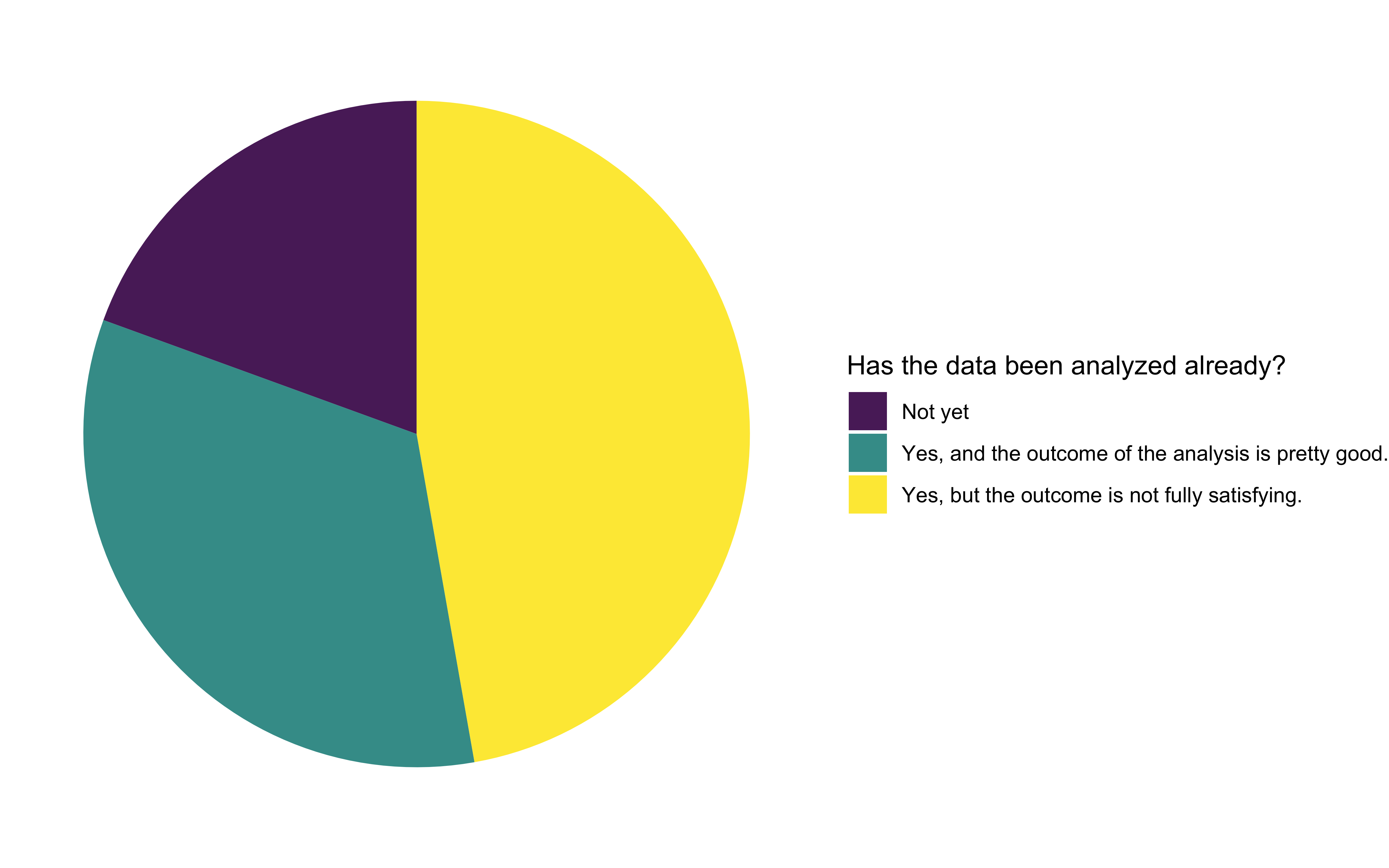
How have applicants addressed their analysis problem so far?
We were pleased to see that most of the applicants had already analyzed the data, but half of them were not fully satisfied with the outcomes. We interpreted this as an opportunity to improve existing workflows. The other half of the applicants were satisfied with their analysis results, but longed for better automation of their workflow, so it becomes less cumbersome and time-consuming. Around 20% of all applications haven’t yet started analyzing their data.
When asked about the tools applicants used to analyze their image data, Fiji and ImageJ were the most frequently used ones. Custom code in Python and Matlab is also popular. Other frequently used tools included Napari, Amira, Qupath, CellProfiler, ilastik, Imaris, Cellpose, and Zen.

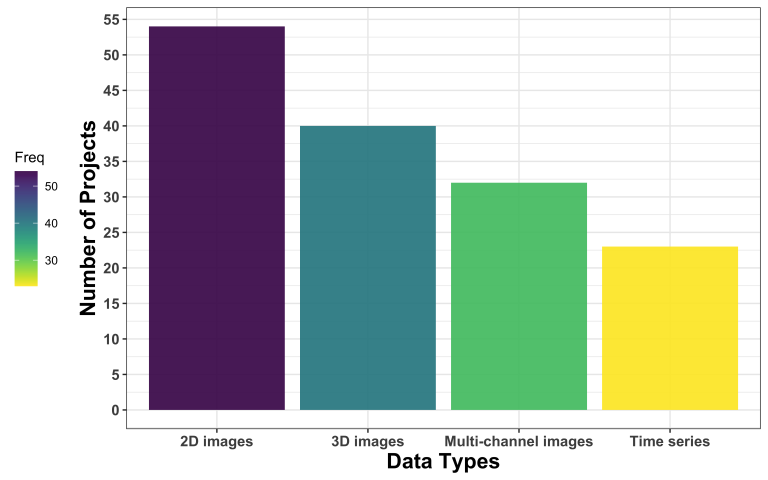
What kind of data and what format do applicants deal with?
The most common kind of image data are 2D images, followed by 3D images, multi-channel images, and time series.
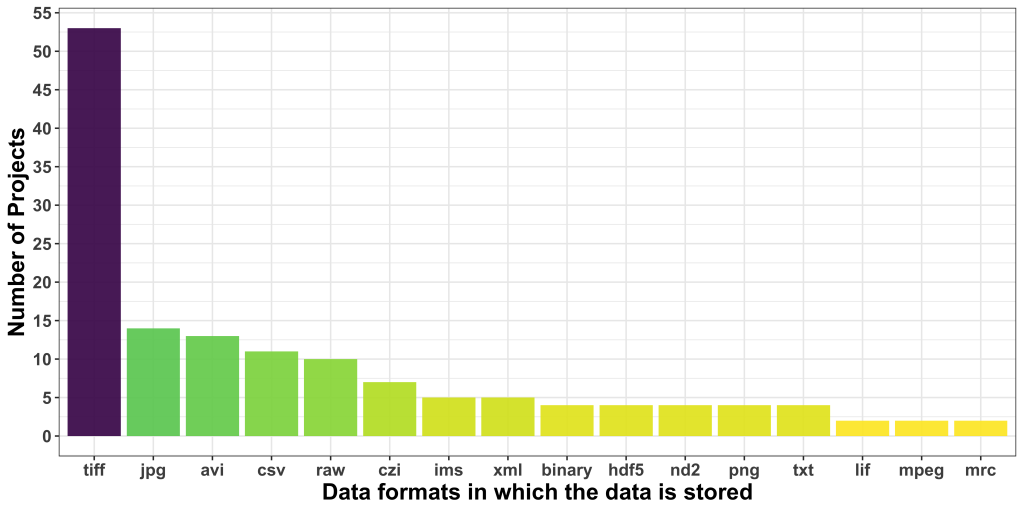
Regarding data formats, TIFF was the most popular one, followed by JPG, which is borderline alarming due to the lossy nature of this format. AVI was the third most common format users seem to be dealing with. CSV was, interestingly, the most common non-image data format.
Additionally, we asked about the relevance of the metadata to address the proposed project. 17 applicants didn’t reply, and 24 others did, but do not think metadata is quite relevant to the problem at hand. While this is likely true for the problem at hand, these responses show that the reusability and FAIRness of acquired and analyzed image data is not yet part of the default mindset of applicants to our Open Call.
How much project-relevant data do the applicants bring?
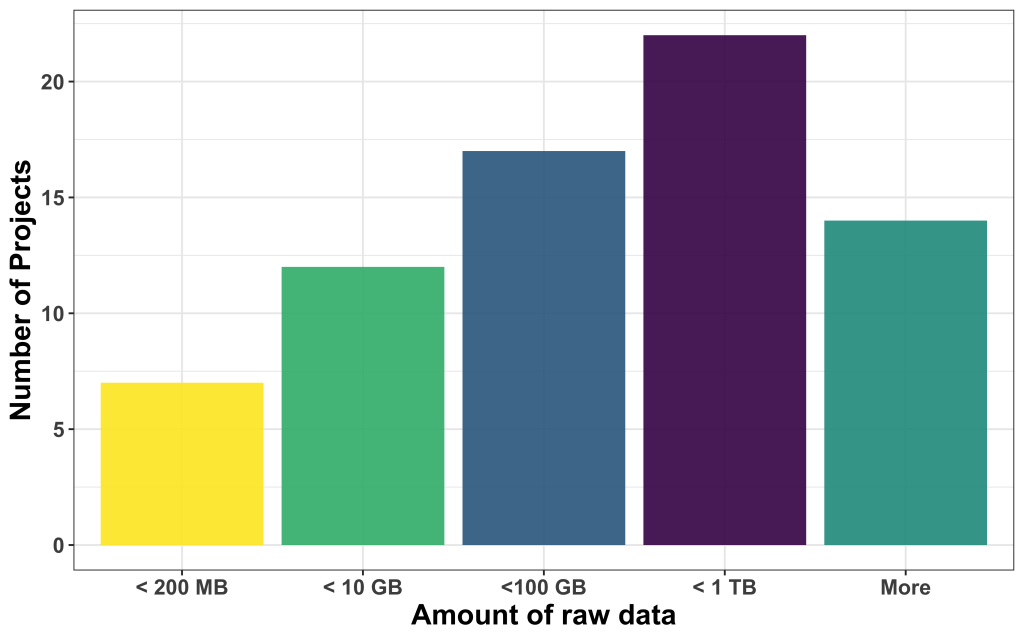
We found that most of the data available was large, with most of them in the range between 100 and 1000 GB, followed by projects that come with less than 10 GB, between 10 and 100 GB, and less than 200 MB. 14 projects had more data than 1 TB to offer.
Is ground truth readily available?
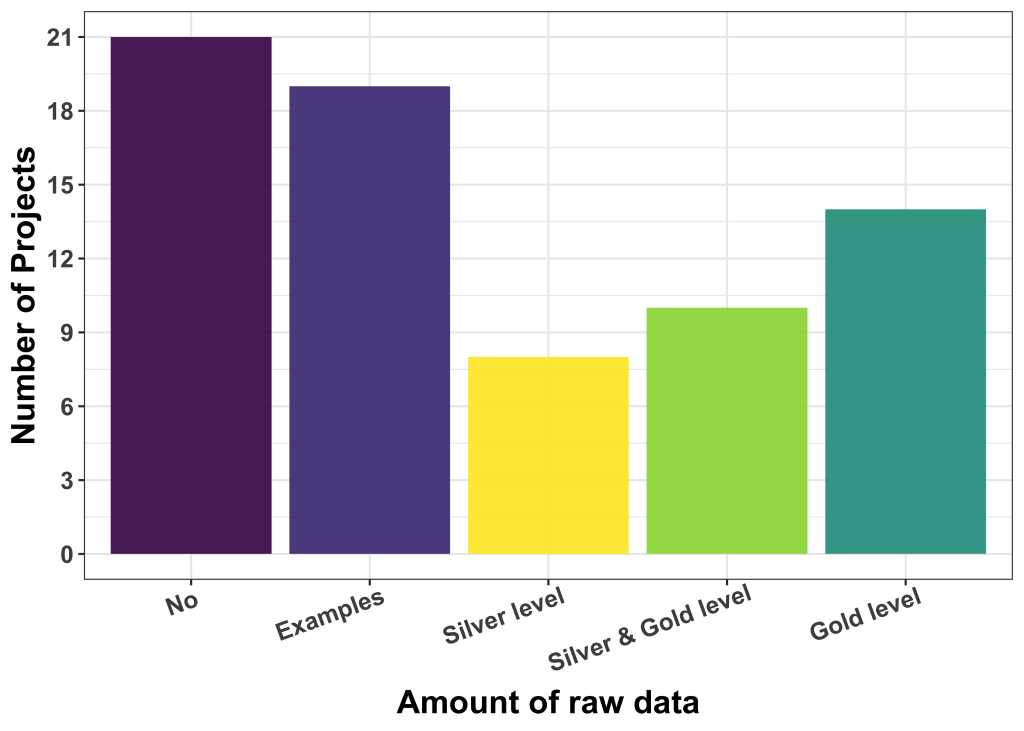
We also asked about the availability of labelled data and provided some guidelines regarding the kind and quality of such labels. We distinguished: (i) Silver ground truth, i.e. results/labels good enough to be used for publication (but maybe fully or partly machine-generated, and (ii) gold ground truth, i.e. human curated labels of high fidelity and quality.
The majority of applicants (40) had no labelled data or only very few examples. The rest have silver level (8), a mix of silver and gold levels (10) and gold level (14) ground truth available. The de-facto quality of available label data is, at this point in time, not easy to be assessed, but our experience is that users who believe to have gold-level label data are not always right.
Is the available data openly shareable?
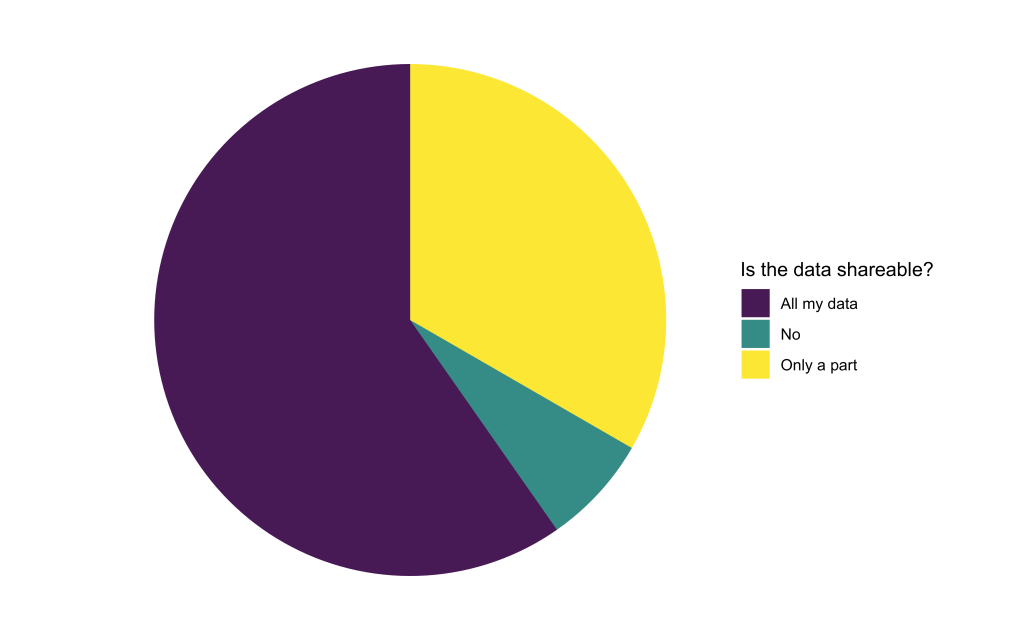
To train Deep Learning models, the computational experts will need access to the available image data. We found that only a small portion of applicants were not able to share their data at all. The rest is willing to share either all or at least some part of their data. When only parts of the data are sharable, reasons were often related to data privacy issues or concerns about sharing unpublished data.
What’s next? How will we proceed?
We are currently undergoing an eligibility check and the pool of reviewers will start looking at the projects in more detail. In particular, they will rank the projects based on the following criteria:
- The proposed project is amenable to Deep Learning methods/approaches/tools.
- Does the project have well-defined goals (and are those goals the correct ones)?
- A complete solution to the proposed project will require additional classical routines to be developed.
- The project, once completed, will be useful for a broader scientific user base.
- The project will likely require the generation of significant amounts of training data.
- This project likely boils down to finding and using the right (existing) tool.
- Approaches/scripts/models developed to solve this project will likely be reusable for other, similar projects.
- The project, once completed, will be interesting to computational researchers (e.g. within a public challenge).
- The applicant(s) might have a problematic attitude about sharing their data.
- Data looks as if the proposed project might be feasible (results good enough to make users happy).
- Do you expect that we can (within reasonable effort) improve on the existing analysis pipeline?
The reviewers will also identify the parts of the project that can be improved, evaluate if deep learning can be of help and provide an estimation of the time needed to support the project.
We will keep you posted about all developments. Thanks for reading thus far! 🙂





















