BioEngine: Unveiling cloud-powered AI for simplified Bioimage Analysis
by Jeremy Metz, Beatriz Serrano-Solano and Wei Ouyang
In a major step toward democratizing AI in life sciences, the AI4Life consortium announces the launch of BioEngine—a scalable, cloud-based infrastructure that powers the BioImage Model Zoo. Designed to be accessible to both experts and novices, BioEngine aims to revolutionize the way bioimage analysis is conducted.
The challenge
The escalating growth of data in life sciences has revealed the limitations of conventional desktop applications used for bioimage analysis. These local solutions are increasingly inadequate to handle high-throughput data and sophisticated applications like AI-driven image analysis. Users often face challenges with large data sets, complex hardware requirements, and intricate software dependencies that make these tools cumbersome to use and difficult to deploy. Additionally, existing machine learning model zoos often necessitate a level of expertise in programming and model selection, making them inaccessible to a wider audience. All these challenges collectively point to the need for a more efficient, scalable, and user-friendly solution for AI model serving.
The solution
Enter BioEngine, a state-of-the-art cloud infrastructure designed to simplify the complex landscape of bioimage analysis. BioEngine powers the BioImage Model Zoo, allowing users to test-run pre-trained AI models on their own images without requiring any local installation. Its cloud-based approach means you can easily connect BioEngine to existing software platforms like Fiji, Icy, and napari, thereby eliminating the need to install multiple dependencies.
For the developers in our community, BioEngine serves as a cloud platform designed to cater to many users while judiciously using limited GPU resources. It features a simple API that can be accessed via HTTP or WebSocket, offering a seamless experience for running models in the cloud. This API can be effortlessly integrated into Python scripts, Jupyter notebooks, or web-based applications.
Key Features
- User-friendly API for quick integration into existing workflows.
- Diverse array of supported model formats including TensorFlow, Torch, and ONNX.
- Cloud-based system for simplifying complex setups, ideal for connecting with existing bioimaging software.
- Upcoming toolkit for on-premise deployment, providing versatile installation options from Kubernetes clusters to individual workstations or laptops.
By employing BioEngine, both experts and non-experts can overcome the challenges associated with traditional desktop-based solutions and enter an era of streamlined, accessible and scalable bioimage analysis.
Technical challenges
Efficiently serving a variety of models on limited GPU resources required the development of a unique framework that can manage dynamic model execution and scheduling.
Our approach
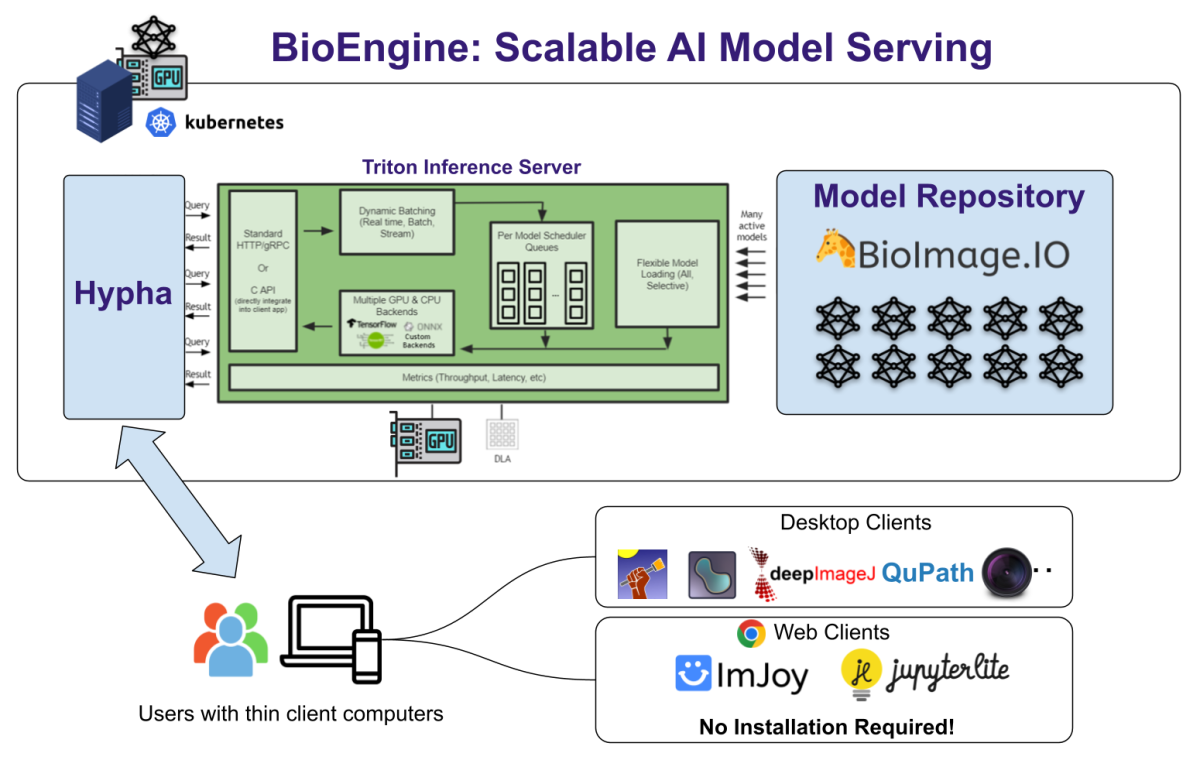
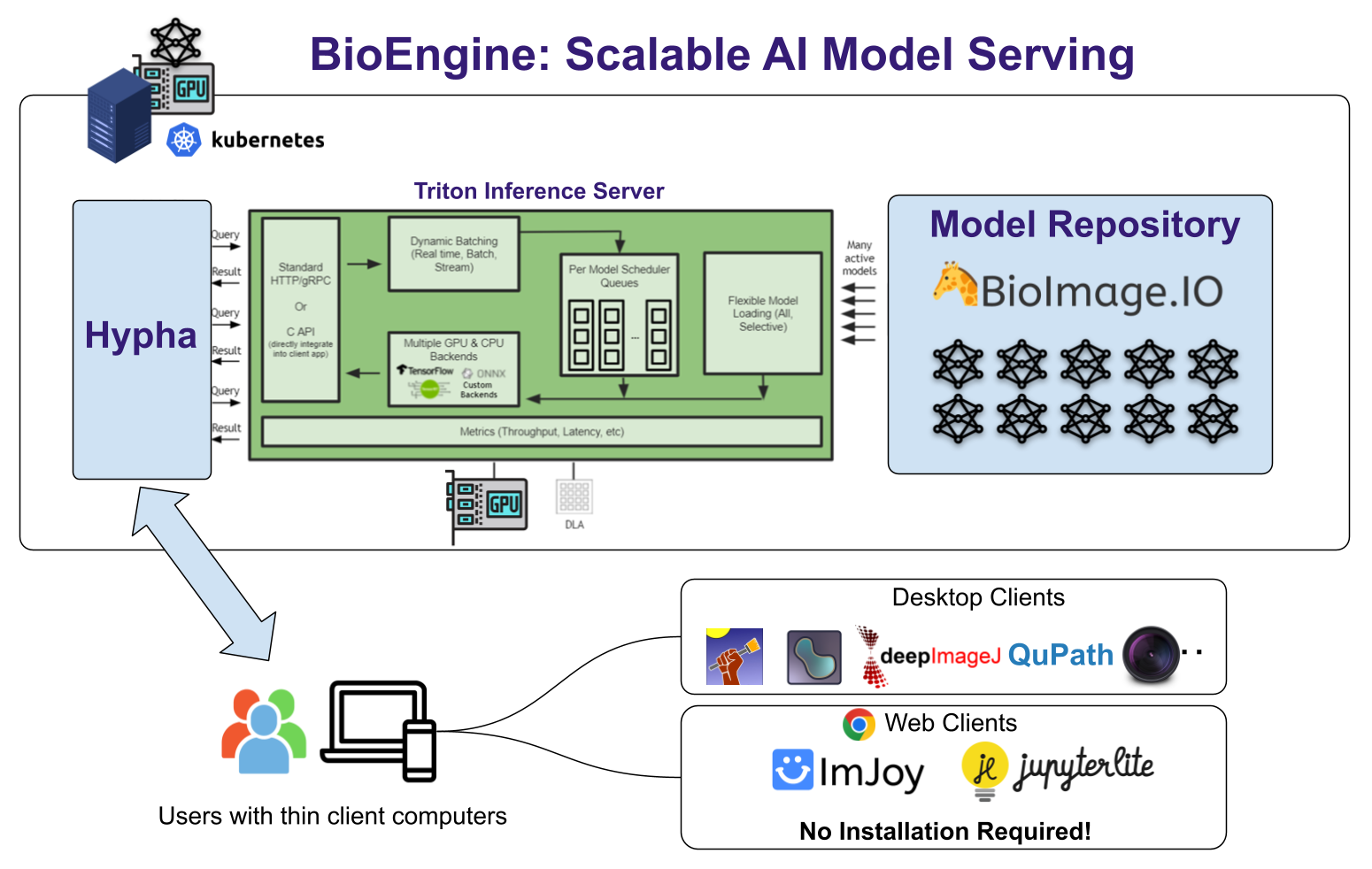
BioEngine is the result of a concerted collaboration primarily between KTH Royal Institute of Technology and the BioImage Archive Team at EMBL-EBI, under the auspices of the AI4Life consortium. The platform is built as an extension of the Hypha framework, a robust RPC-based communication hub that orchestrates containerized components for a seamless and efficient operation.
Technical Underpinnings
- Hypha Framework: The bedrock of our BioEngine, Hypha facilitates the orchestration of containerized modules, enabling a robust system architecture. It supports key functionalities like user authentication and virtual workspaces.
- Nvidia Triton Inference Server: For handling AI models, BioEngine leverages the Nvidia Triton Inference Server. This allows us to manage and dynamically schedule GPU workloads based on incoming user requests, ensuring optimal resource allocation and efficiency.
- S3-Based Object Storage: All data is securely and efficiently stored using S3 object storage, ensuring both speed and reliability.
Developer-friendly API
BioEngine provides both HTTP and WebSocket-based RPC APIs. Developers can effortlessly integrate these APIs into web and desktop apps, thereby streamlining the interaction between software and the cloud-based AI models.
Key Features
- Simple API for model execution
- Scalability across various AI frameworks like TensorFlow, Torch, and ONNX
- Support for multiple types of image analysis tasks
- A variety of data storage and retrieval options
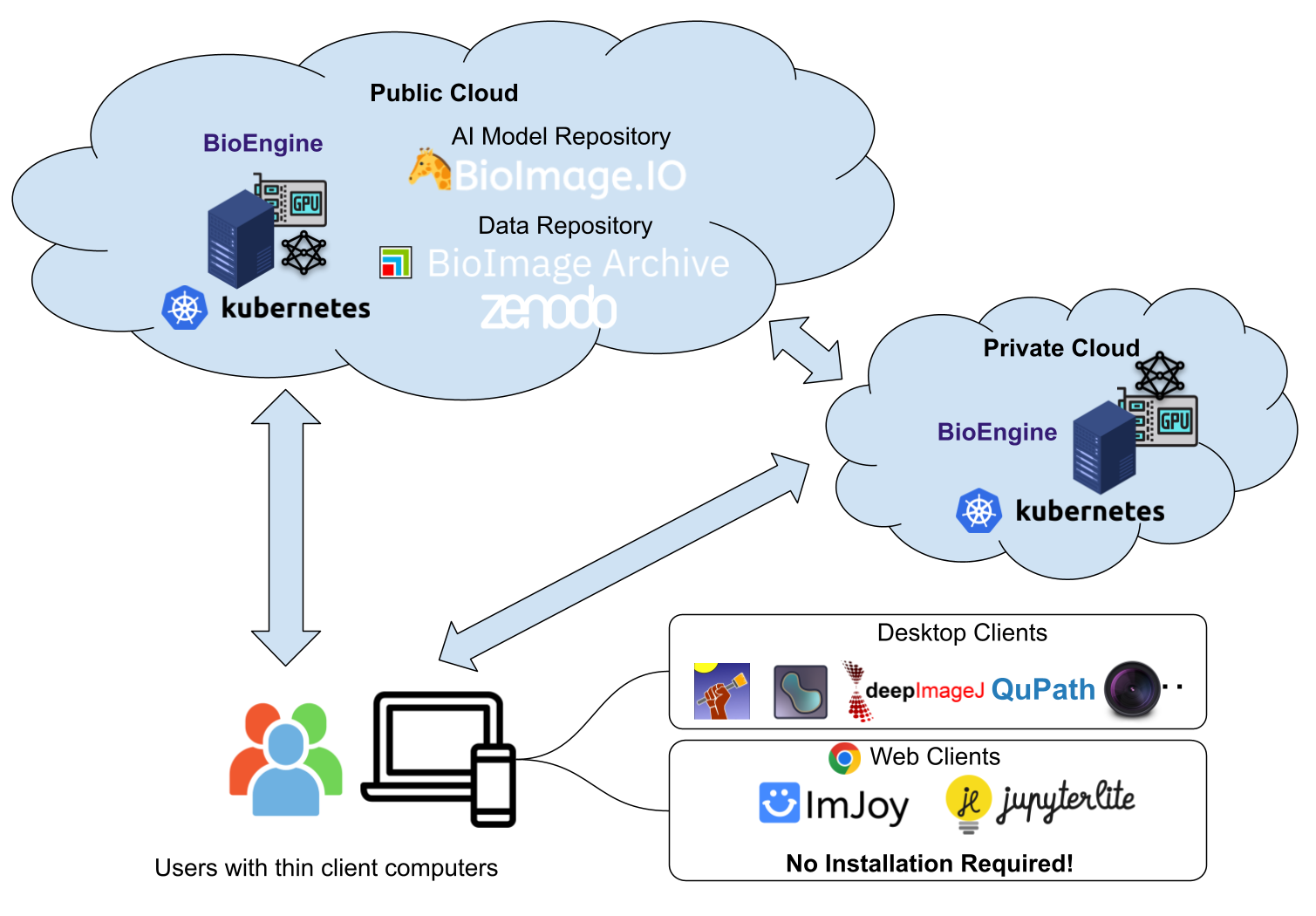
Deployment Options
We are actively working on broadening our deployment offerings:
- Kubernetes (K8s) Cluster: For large-scale, enterprise-level applications, we are developing support for auto-scaling within a K8s cluster environment.
- Docker-Compose: For smaller-scale or local deployments, Docker-Compose options are also in the pipeline.
Cost and Accessibility
With the support the EU Horizon research infrastructure grant, we are committed to promoting accessibility and lowering barriers to advanced bioimage analysis:
- Free and Open-Source: For testing and evaluating, BioEngine will offer a free and open-source option.
- On-Premise Deployment: For institutions seeking to integrate BioEngine into their existing IT infrastructures, we are developing versatile on-premise deployment solutions.
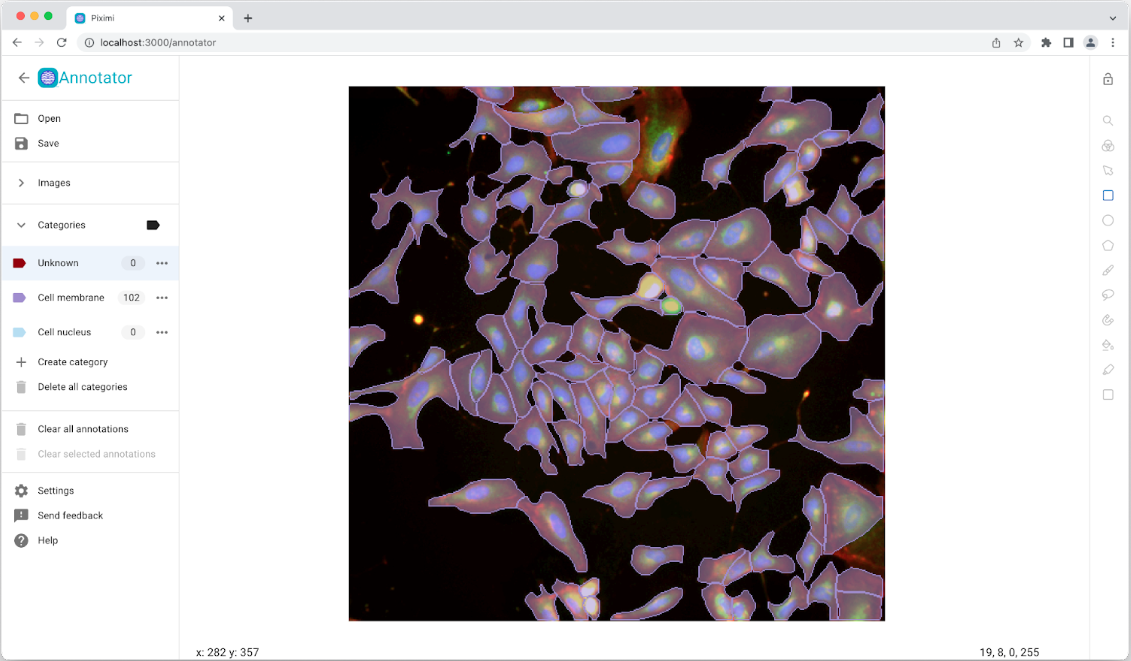
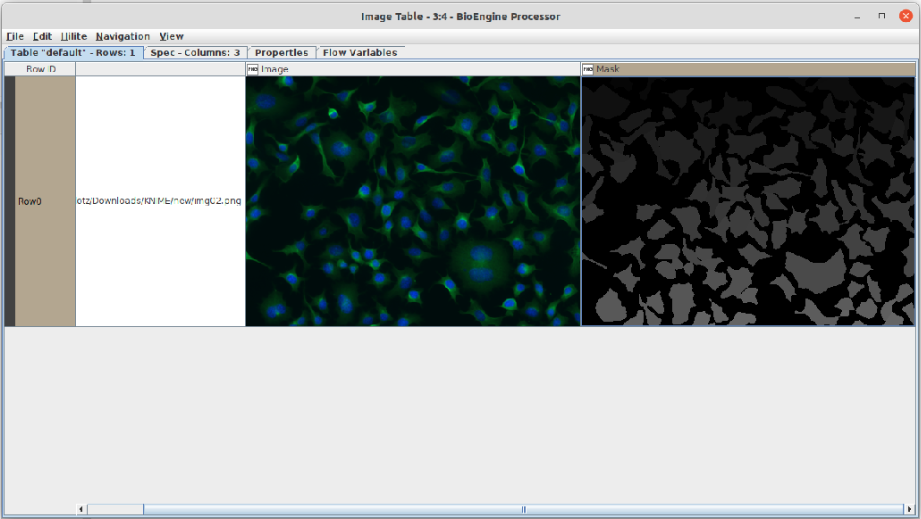
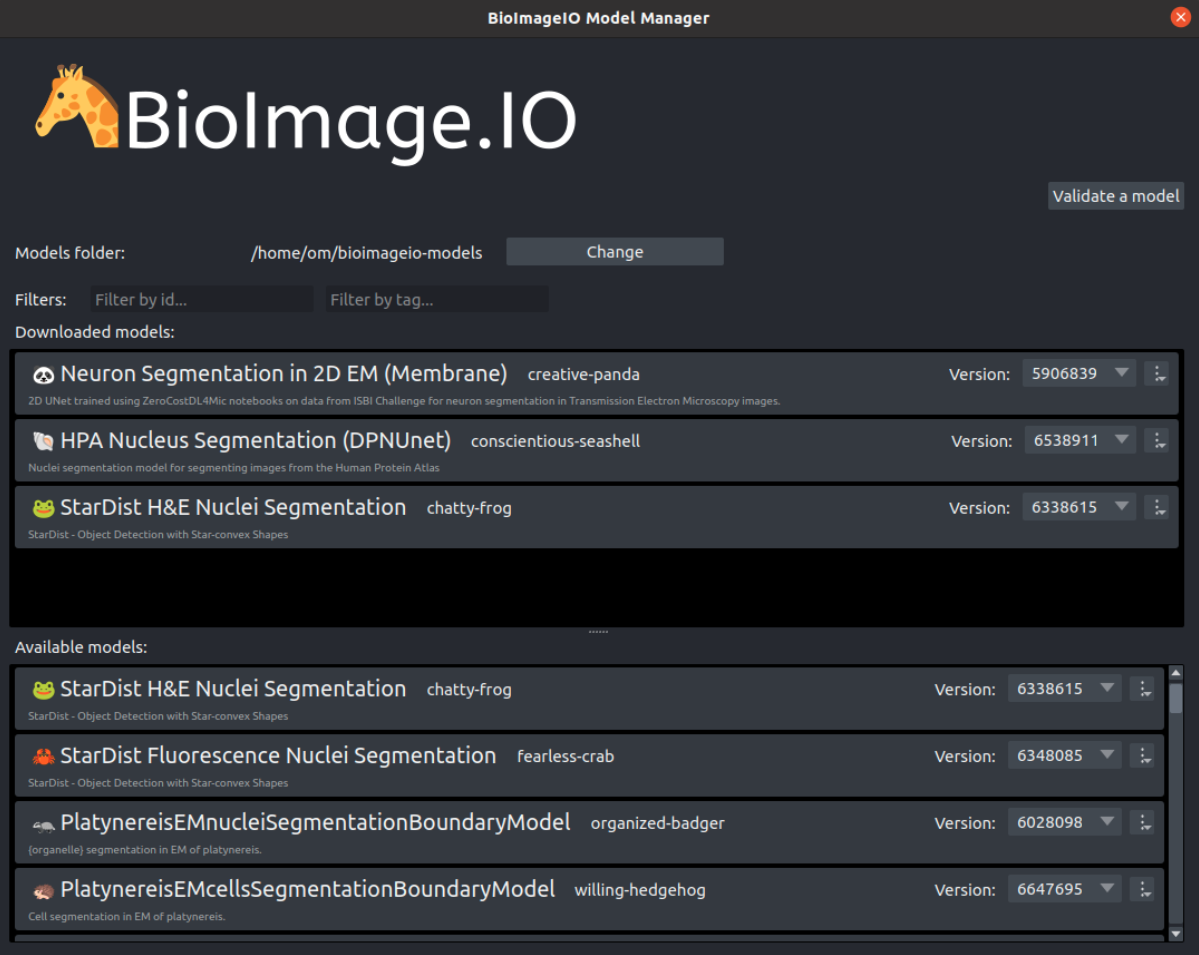
BioEngine integration examples
Invitation to the community
We encourage everyone to try BioEngine and provide feedback (via image.sc forum, GitHub issues, or our contact form). Your input is crucial for the continual refinement of this revolutionary platform.
For Developers and Bioimage Analysts
BioEngine offers an easy-to-use API for running models, simplifying software design and deployment.
This API can be integrated into Python scripts, Jupyter notebooks, or web-based applications, giving you the flexibility to adapt BioEngine to your own projects.
Looking ahead
We are actively working on an easily deployable package for institutional use and aim to improve stability and scalability.
For more information, please refer to our API documentation.
With BioEngine, we are democratizing advanced bioimage analysis by offering cloud-based, plug-and-play AI solutions that can be effortlessly integrated into existing software ecosystems, thereby accelerating both research and real-world applications in life sciences.
Acknowledgements
- EU: Funded by the EU’s Horizon Europe program, grant no. 101057970.
- Nvidia: Supported our development, notably with Triton Inference Server and computing credit.
- Bioimaging Community: Thanks to testers and AI4Life Stockholm Hackathon 2023 participants for vital feedback.
- SciLifeLab & KAW: Supported by SciLifeLab DDLS program, funded by Knut and Alice Wallenberg Foundation.
- The authors utilized the language model ChatGPT developed by OpenAI for assistance in the structuring and drafting of this news article.