DL4MicEverywhere: reproducible and portable deep learning workflows for bioimage analysis
by Estibaliz Gómez-de-Mariscal, Iván Hidalgo-Cenalmor, Mariana G Ferreira, Ricardo Henriques
DL4MicEverywhere (https://github.com/HenriquesLab/DL4MicEverywhere) is a platform developed within the AI4Life project. It offers researchers an easy-to-use gateway to reproducible and portable deep learning techniques for bioimage analysis. The platform utilizes Docker containers to encapsulate deep learning-based approaches together with user-friendly interactive notebooks, guaranteeing smooth operation across various computing environments such as personal devices or high-performance computing (HPC) systems1. It currently incorporates numerous pre-existing ZeroCostDL4Mic notebooks –yet another community partner– for tasks such as segmentation, reconstruction, image translation and image generation.

The functionalities of DL4MicEverywhere are supported by a user-friendly GUI that allows users to rapidly launch the Docker containers and interact with the notebooks in a zero-code fashion. This interface is designed in a way that public methods available in the BioImage Model Zoo or local ones can be automatically launched without dealing with the intricacies of Docker configurations, environment setups, or coding. This is enabled by the advanced mode of the user interface.
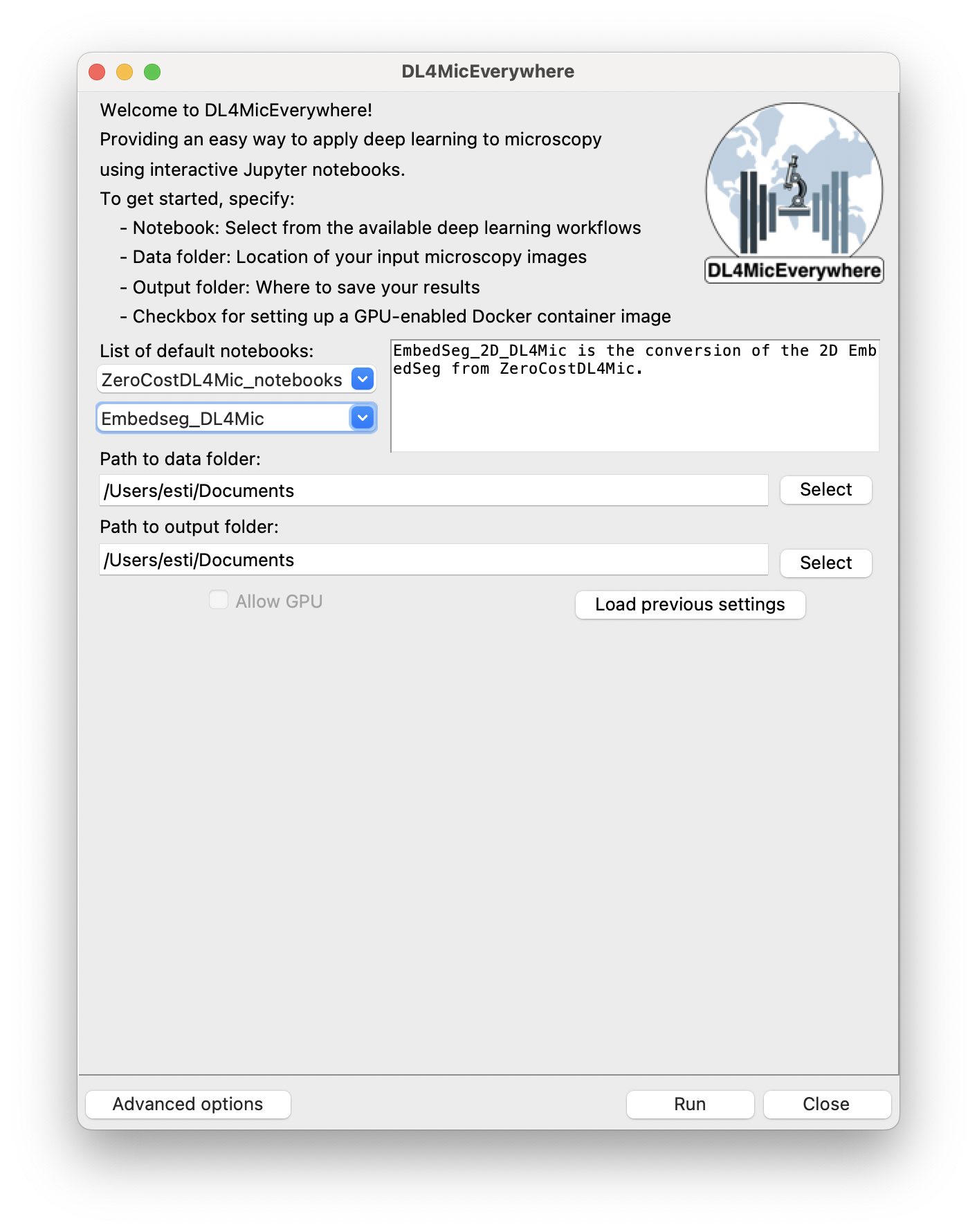
a)
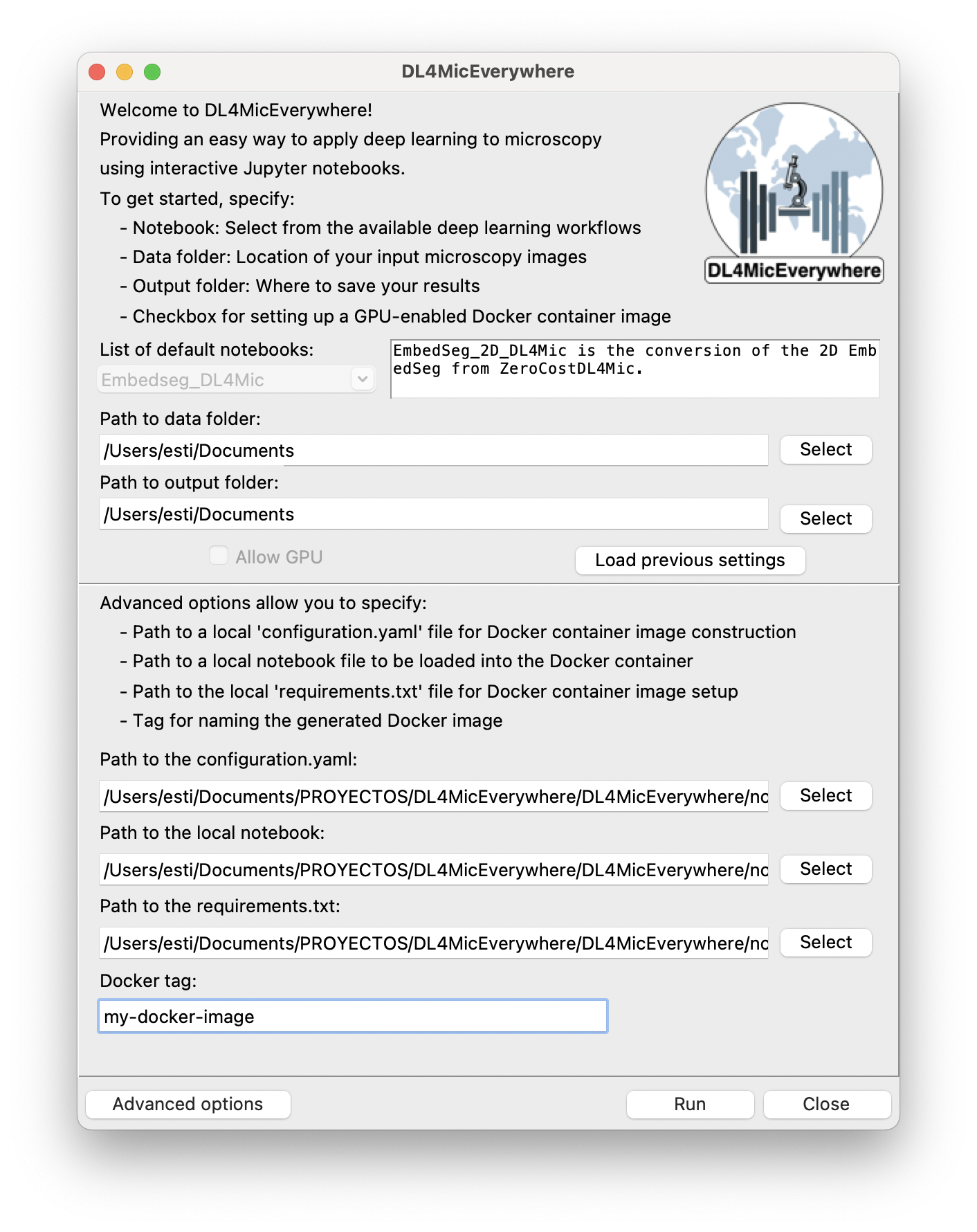
b)
Figure 1: DL4MicEverywhere interface models. a) The basic mode allows the launch of containerised notebooks that are publicly available and tested. b) Advanced options allowing for the automatic containerisation of local or private models.
Watch a brief tutorial: https://youtu.be/rUt1aG_AXh8?si=p8cbcaWU2RWTE1mf
Read the preprint: I. Hidalgo-Cenalmor et al., DL4MicEverywhere: Deep learning for microscopy made flexible, shareable, and reproducible, bioRxiv 2023, https://doi.org/10.1101/2023.11.19.567606
Empowering Developers
DL4MicEverywhere serves as an infrastructure and service for containerising deep learning methods in the context of bioimage analysis. The platform provides the tools to automatically containerise their methods and ensure the correct configuration of the built Docker images.
These are the key features of DL4MicEverywhere:
- Automatic containerisation of the BioImage Analysis pipelines. The platform automatically builds Docker images for AMD64 (Windows/Linux/macOS – Intel) (with and without GPU access) and ARM64 (macOS-M1/M2) systems. This process controls the versions of the required libraries upstream and downstream of the Docker container, enabling the automatic containerisation of bioimage analysis pipelines.
- Integration of user-friendly Jupyter Notebooks: It allows the encapsulation of Jupyter Notebooks for high-level and documented programmatic interaction. These notebooks can be automatically converted into interactive interfaces for a zero-code experience.
- Continuous integration system: DL4MicEverywhere incorporates an automatic validation pipeline to test the correct containerisation of image processing pipelines. This ensures the reliability and accuracy of the containerised methods, contributing to their robustness and reproducibility.
- Publicly available Docker images in Docker Hub (https://hub.docker.com/r/henriqueslab/dl4miceverywhere/): The platform automatically uploads validated Docker images to Docker Hub, ensuring their long-term accessibility.
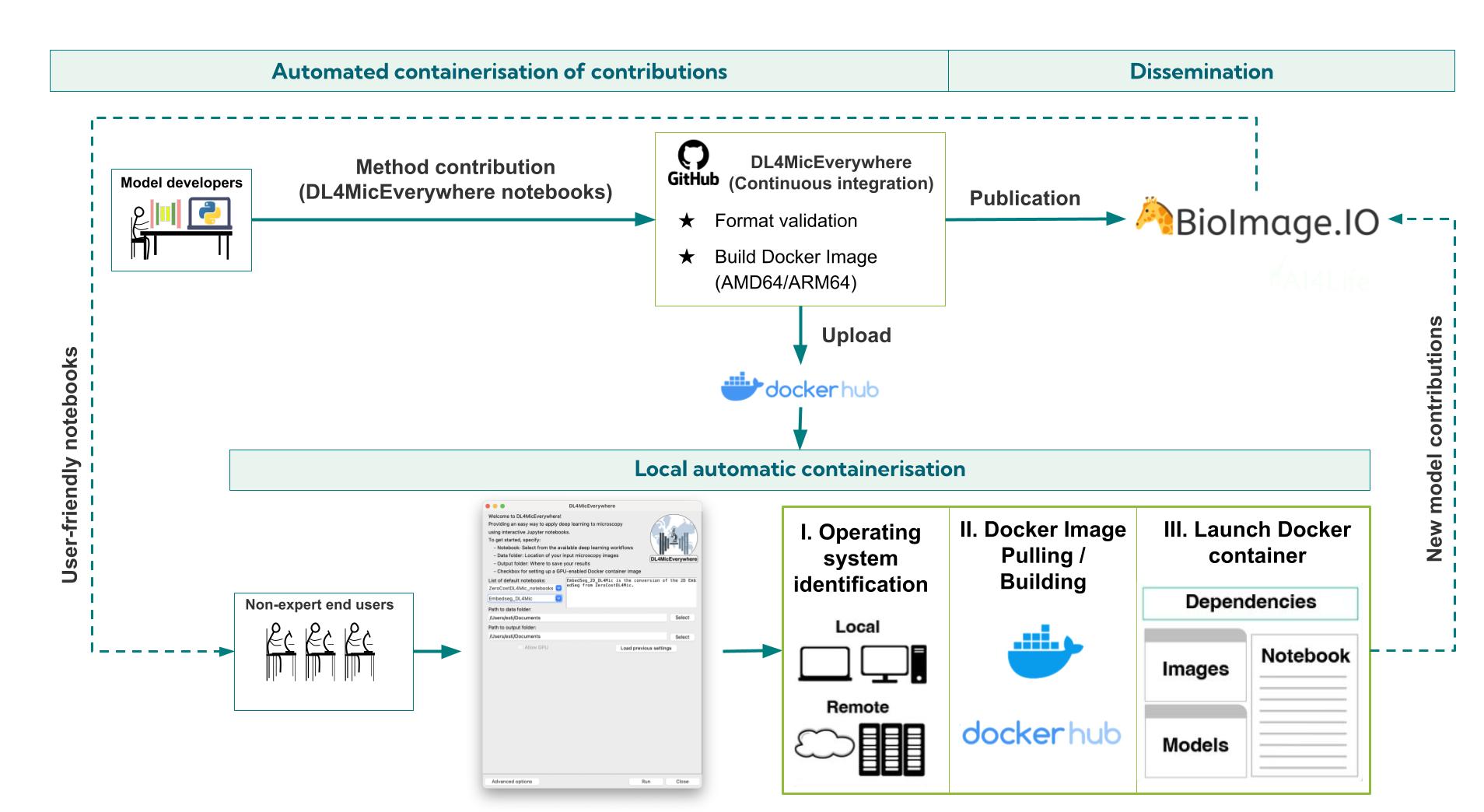
Figure 2: Schematic description of the automatic containerisation proposed by DL4MicEverywhere. Developers can contribute their models within DL4MicEverywhere notebooks directly to the DL4MicEverywhere GitHub Repository. The repository runs an automatic continuous integration pipeline to test the format of the notebooks, the correct building of Docker Images and publishes a versioned Docker Image in Docker Hub. This containerisation is synchronised with the BioImage Model Zoo and follows the same specifications, ensuring that the methods are accessible to non-expert users. Non-expert users access the containerised workflows with a user-friendly graphical user interface (GUI) that automatically launches the Docker container corresponding to the operating system and configuration of the users. Once the Docker container is set up, the users can interact with the method directly in Jupyter Notebooks without dealing with the intricacies of Docker containerisation. Likewise, the users will be able to reproduce the pipelines, train their models and contribute them to the BioImage Model Zoo within a reproducible and portable ecosystem.
1 Docker containers allow the full virtualisation of computational environments without affecting local installations. They allow building the specific environments and dependency setup needed for each workflow. These virtualisations, once built, they are portable and installable across systems. Therefore, they are highly recommended to ensure the reproducibility of computational pipelines.

