AI4Life Standards and Interoperability
by Teresa Zulueta-Coarasa, Fynn Beuttenmueller, Anna Kreshuk, Beatriz Serrano-Solano
In AI4Life, we believe that interoperability and standardisation are the backbone of a healthy AI research ecosystem, allowing data and models to be reused and combined across different research groups, institutions, and platforms. Without standards, valuable datasets and AI models often remain underutilised, difficult to reuse, reproduce, and less impactful than they could be.
One of the main goals of AI4Life has been to create and promote standards for sharing AI models and AI-ready datasets for biological images. By doing this, we aim to ensure that data and models are truly FAIR (Findable, Accessible, Interoperable, and Reusable) so they can support scientific discovery for years to come.
Setting standards for biological image datasets
In January 2023, the BioImage Archive organised a workshop that brought together 45 experts from diverse backgrounds: data producers, annotators, curators, AI researchers, bioimage analysts, and software developers. Together, they defined recommendations for sharing annotated, AI-ready biological image datasets.
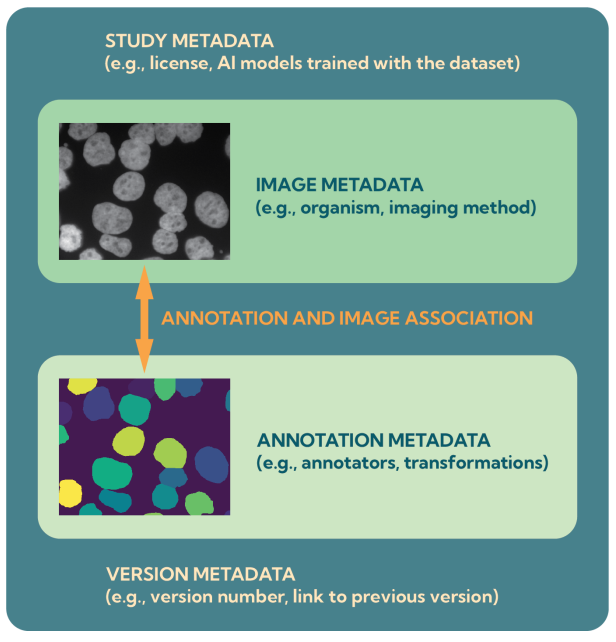
These recommendations are grouped under the acronym MIFA:
- Metadata: clear information about datasets and annotations.
- Incentives: giving proper recognition to dataset creators.
- Formats: adopting a small set of interoperable formats, such as OME-Zarr.
- Accessibility: making datasets openly available in repositories like the BioImage Archive.
The MIFA guidelines have been published in Nature Methods (https://www.nature.com/articles/s41592-025-02835-8). They are expected to help researchers more easily train and evaluate AI models across diverse biological imaging tasks and unlock the value of archived imaging data.
A standard for AI models
In addition to datasets, AI4Life also supports a model metadata standard. This standard describes how pre-trained models should be documented so that others can find, reuse, and integrate them into their work. It is openly available and registered in FAIRsharing, a trusted global resource for standards, repositories, and policies.
The model standard is implemented through the bioimageio.spec Python package, which provides a versioned metadata format for models. It works with the bioimageio.core library offers utilities and adapters to make models compatible with different tools and frameworks.
With this approach, models can be shared in a way that is:
- Findable: authors and citations are clearly tracked.
- Accessible: models and their documentation are available through the bioimage.io website.
- Interoperable: the model metadata allows for programmatic execution through the bioimageio.core library makes models seamlessly usable through all our Community Partner tools or simply through Python or Java code.
- Reusable: thanks to the metadata, model inference can be executed in a standardised way even without access to the model’s original code.
The BioImage Archive has developed the AI4Life Model Evaluation Platform to benchmark datasets and models more directly, building bridges between the BioImage Archive and the BioImage Model Zoo.
While pre-trained models are already very useful, they are even more powerful when bundled together with their training datasets and training code. Model metadata supports linking to datasets and code by introducing the corresponding metadata field and a minimal description format for datasets and notebooks.
The dataset description is currently available in bioimageio.spec serves as a starting point; plans are underway to extend this with deeper integration of the MIFA guidelines. In the future, this will make programmatic access to well-described datasets even easier, enabling researchers worldwide to train, compare, and improve AI models for bioimaging.
