Expanding AI4Life Training Resources: New and Updated Materials
by Caterina Fuster-Barceló
To enhance accessibility and usability, the European consortium AI4Life has expanded and updated the BioImage Model Zoo (BMZ, BioImage.IO) training materials as part of the D3.3 Deliverable (available in zenodo). This effort involved evaluating existing resources, introducing new materials, and updating documentation to ensure comprehensive guidance for both developers and end-users of the BMZ.
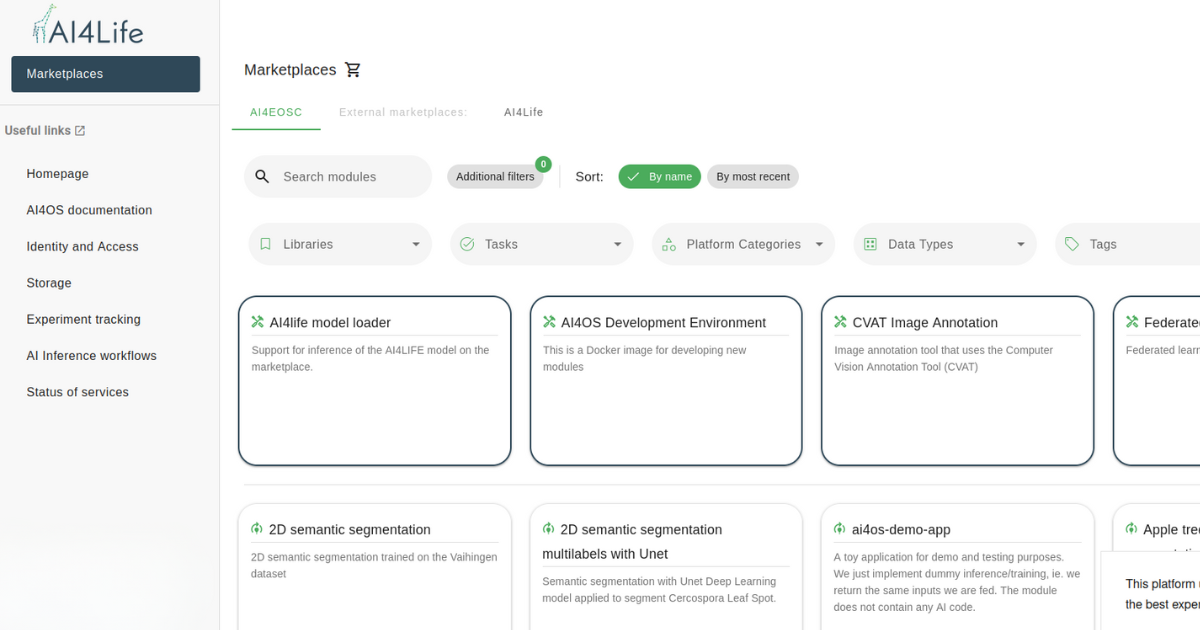
Currently, the AI4Life training material list includes over 30 different training materials, spanning website documentation, case studies, video tutorials, computational notebooks, and slides. The majority of resources are targeted at end-users (47.1%) and developers (32.4%), with additional materials designed to serve both audiences (20.6%). The updated distribution of training materials and their target audiences is visualized in the figures below.


Key Insights from the Updated Training Materials
- Video tutorials (14) remain the most used format, followed by website documentation (9) and computational notebooks (6).
- Nearly half of the training materials cater to end-users, helping them navigate BMZ and integrate AI models into workflows.
Training Documentation Aligned with the AI Model Lifecycle
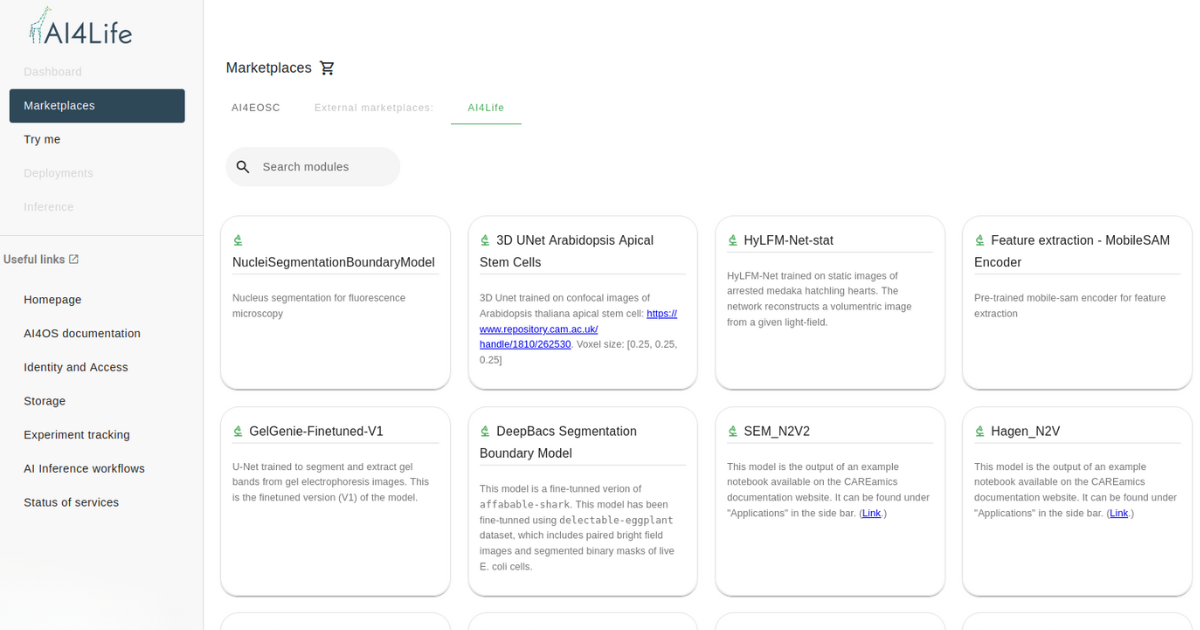
To ensure that documentation is structured in a way that is intuitive and practical for users, we have aligned its training materials with the lifecycle of a Deep Learning (DL) model in the BMZ. The image below illustrates this workflow-based documentation approach, categorizing resources according to the key stages of models and the targeted users of each case.
Each stage of the DL model lifecycle—model development, packaging, upload, accessibility, deployment, and fine-tuning—is supported by a combination of website documentation, video tutorials, computational notebooks, and case studies. Developer-focused documentation (highlighted in orange) ensures that DL models are properly packaged, documented, and submitted to BMZ, while end-user documentation (highlighted in blue) provides guidance on how to access and integrate models into image analysis workflows.

Highlighted Updates and New Materials
1. New Guided Tutorials on YouTube. AI4Life has added three new step-by-step video tutorials to help users and developers navigate BMZ more efficiently.
2. Comprehensive Guide on Running BMZ Models in Community Partner Tools. A new documentation resource provides clear, step-by-step instructions on how to execute BMZ models across different bioimage analysis software.
3. Expanded Documentation on the Resource Description File (RDF). To improve model documentation clarity, AI4Life has introduced updated materials explaining the BMZ Resource Description File (RDF) in various formats, making it more accessible to different user groups.
These updates strengthen AI4Life’s commitment to providing high-quality, accessible training materials that support the growth and usability of the BioImage Model Zoo. By continuously refining and expanding these resources, AI4Life ensures that bioimage analysis researchers and developers have the tools they need to efficiently integrate, deploy, and utilize AI models in their scientific workflows.