Welcome to the AI4Life Denoising Challenge 2024!
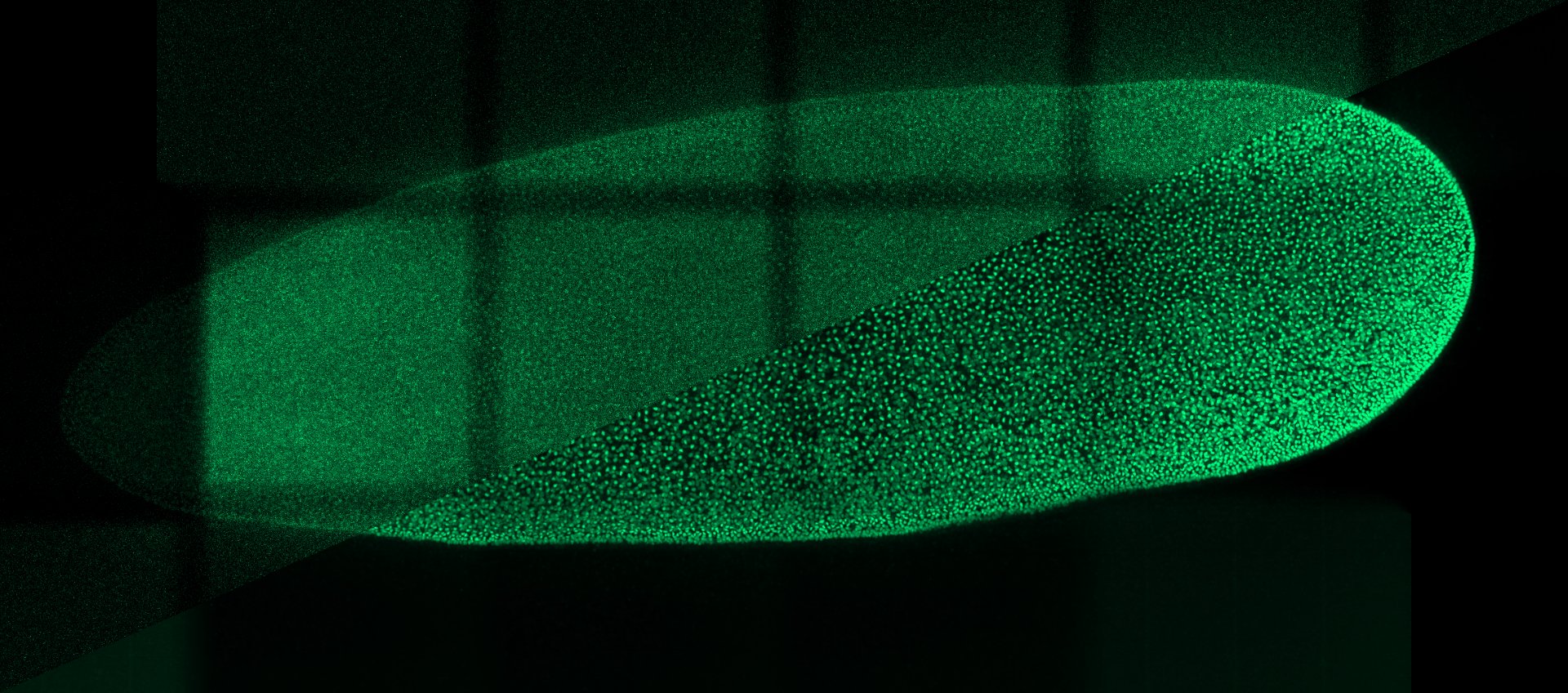
Weigert, M., Schmidt, U., Boothe, T. et al. Content-aware image restoration: pushing the limits of fluorescence microscopy. Nat Methods 15, 1090–1097 (2018)
Biological motivation
Microscopy images are crucial for scientific research, particularly in biology and medicine. However, these images often suffer from various types of noise introduced during the image acquisition process, which can degrade the quality of the images and significantly complicate their interpretation. Denoising microscopy images is therefore essential to improve image quality, all the while preserving important features such as edges, textures, and fine details.
In recent years, deep learning algorithms emerged as a successful approach to removing noise while retaining useful signals. Unlike classical algorithms, which use defined mathematical functions to remove noise, deep learning methods learn to denoise from example data, providing a powerful content-aware approach. Deep learning denoising is already assisting many researchers with the analysis of their acquired images and significantly improves the quality of downstream deep learning approaches such as segmentation or classification.
Aim of the challenge
In this challenge, we want to focus on an unsupervised denoising task, which is particularly challenging and key for the bioimage community. Unlike supervised learning, which requires pairs of noisy and clean images, unsupervised learning is trained using only noisy images, making it a more accessible approach given the difficulty and labour-intensive nature of paired dataset acquisitions.
We invite researchers to apply their deep learning algorithms to four datasets featuring two types of noise: structured and unstructured. We aim to compare the performance of both existing denoising methods and novel ones.
By participating, researchers can contribute to a critical area of scientific research, aiding in interpreting microscopy images and potentially unlocking discoveries in biology and medicine.
Challenge tasks
Unstructured denoising
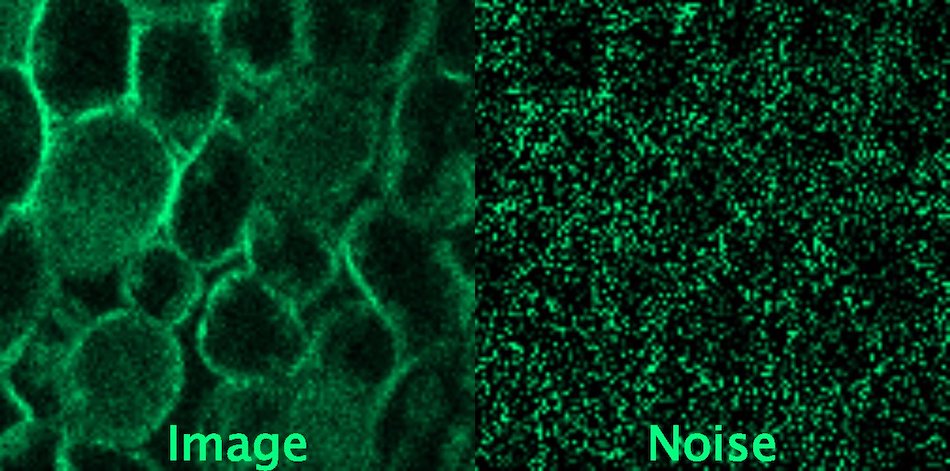
Hagen GM; Bendesky J; Machado R; Nguyen T; Kumar T; Ventura J (2021): Supporting data for “Fluorescence Microscopy Datasets for Training Deep Neural Networks” GigaScience Database
In practice, due to the acquisition process, noise can be highly correlated between neighbouring pixels in various microscopy methods (like confocal microscopy), creating regular patterns in the image.
We invite participants in this challenge to explore existing and novel deep learning denoising approaches using images containing structured noise sampled from two datasets: Fluorescence Microscopy Datasets for Training Deep Neural Networks by Hagen et al., and data from SUPPORT (Statistically unbiased prediction enables accurate denoising of voltage imaging data) method.
Structured denoising
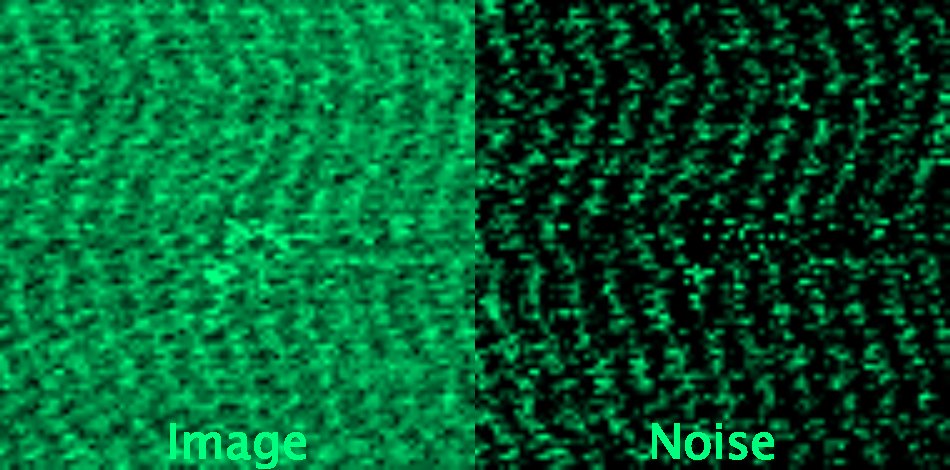
Krull, A., Vicar, T., Prakash, M., Lalit, M., & Jug, F. (2020). Convallaria dataset for microscopy image denoising benchmark as used in Probabilistic Noise2Void paper
Most dominant sources of noise are assumed to be pixel-independent (such as Poisson shot noise and additive Gaussian read-out noise), meaning that neighboring pixels do not carry any information on the amount of noise in one another.
We ask participants to try existing and novel deep learning denoising approaches with images containing unstructured noise sampled from two datasets: JUMP Cell painting Datasets and W2S: Microscopy Data with Joint Denoising and Super-Resolution for Widefield to SIM Mapping.
About this challenge
This challenge is organized by the AI4Life Open Calls and Open Challenges team. We aim to showcase bioimage analysis problems to a broader audience of computational experts and work together in friendly competition, jointly pushing the boundaries of what automated solutions can do for a given analysis task.
How do I participate?
Grand Challenge is hosting this challenge, so please head to
https://ai4life-mdc24.grand-challenge.org/ai4life-mdc24/
and join the fun!



















