Use cases
Explore the real-world projects addressed and selected through our Open Calls, showcasing the diverse applications of AI methods in life sciences.
PROJECT OC1-06
Analysis of the fiber profile of skeletal muscle
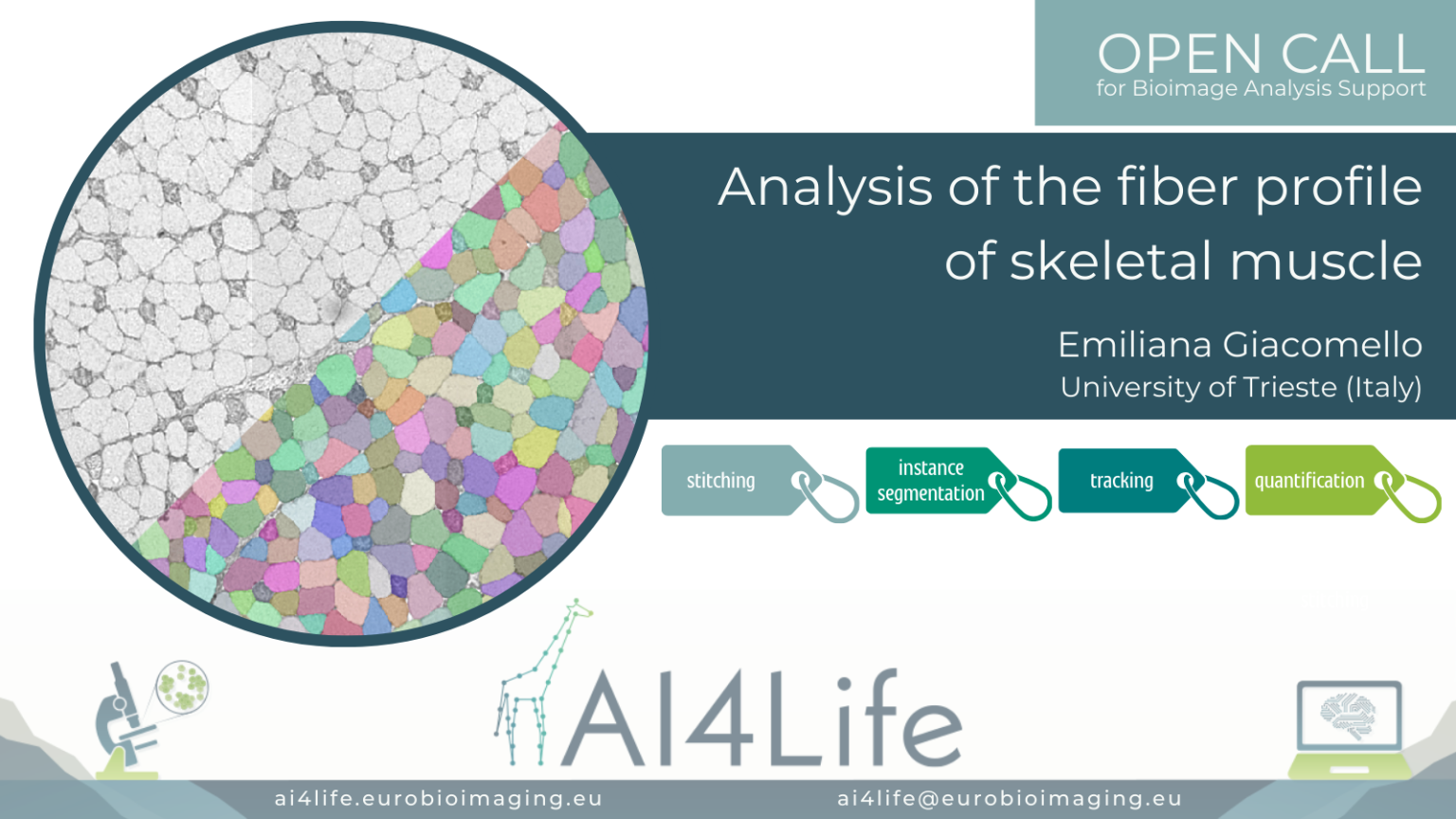
The problem
Researchers from the University of Trieste are studying the regulation of the metabolical properties in murine skeletal muscles by analyzing a series of sections of a single muscle stained with different markers and imaged with light microscopy. The goal of the analysis is to segment all cells in each section and identify them across different acquired images. Although the cells should be present in each section, their morphology can be vastly different, making identification across sections a challenging task.
The solution
As a first step, we aligned the different muscle slices to each other using BigWarp – a Fiji plugin for sample registration and alignment. We then segmented the cells with Cellpose – a popular cell segmentation deep learning library. Finally, to link the resulting cell segmentation across multiple slices, we used the Fiji Trackmate plugin.
PROJECT OC1-66
Automated and integrated cilia profiling
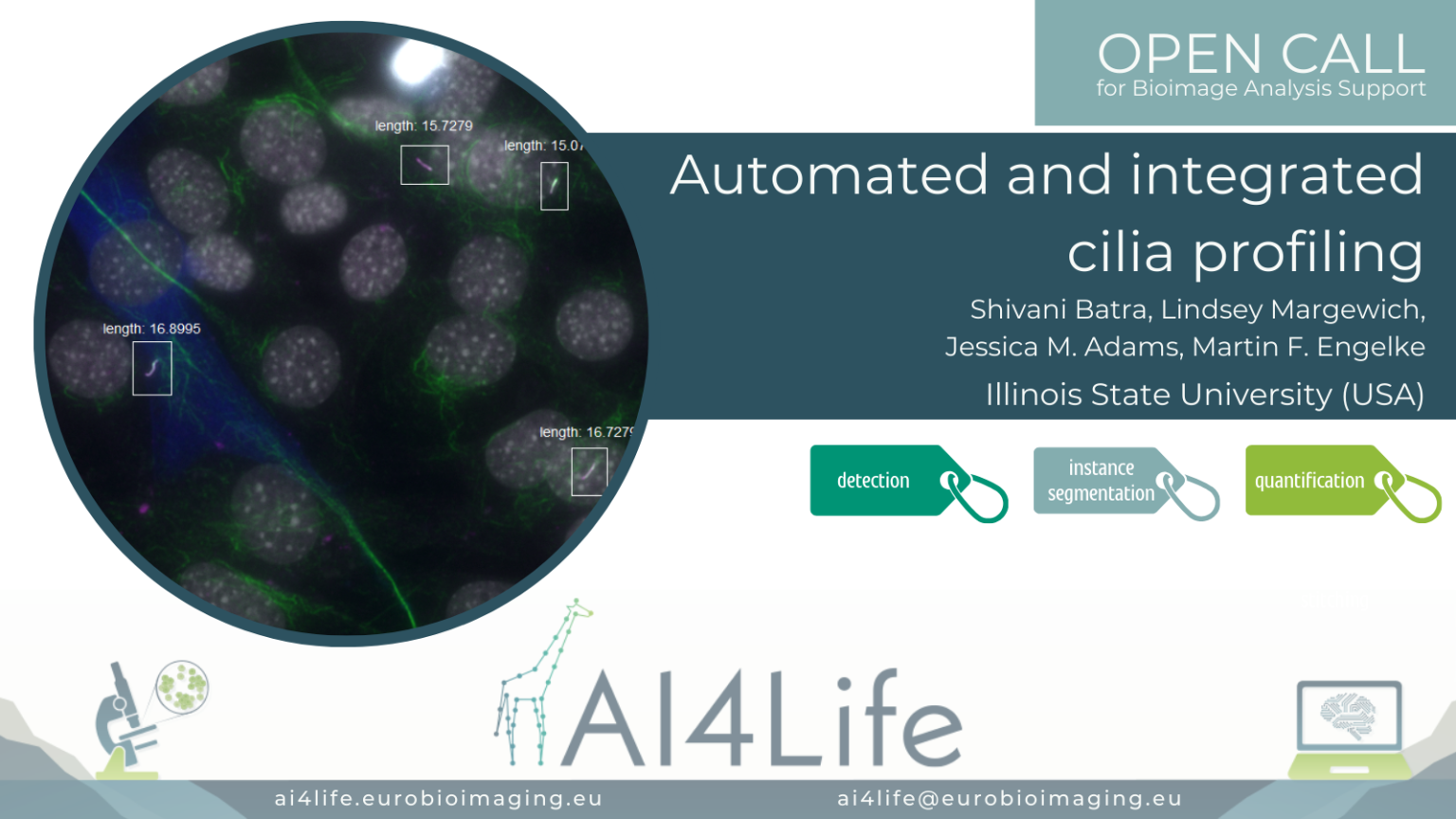
The problem
Researchers at Illinois State University are studying how the expression level of motor transport proteins affects their function in mediating the assembly and length of cilia.
The solution
To do this, they use fluorescence microscopy in conjunction with AI-based image analysis of hundreds of cells. This complex analysis required the integration of three separate segmentation steps: 1) the cell bodies of the cells ectopically expressing fluorescently labelled motor proteins, 2) the nuclei of all of the cells, and 3) the cilia themselves (a challenging task requiring the integration of two separate markers).
The first two segmentations were achieved using a custom deep-learning model trained in Cellpose, while the last was performed by random-forest pixel classification using Labkit. To approximate expression level, the fluorescent intensity of the motor protein was measured in a perinuclear region surrounding the nucleus, while the length of the cilia belonging to expressing and non-expressing cells was measured. These measurements can be combined to correlate cilia length to the intensity of motor protein expression across the entire field of cells.
PROJECT OC1-52
Atlas of microalgae in plankton symbioses revealed by 3D electron microscopy

The problem
In this project, the main challenge faced by the researchers from the CNRS and Grenoble University was the segmentation of various organelles of microalgae in free-living cell and symbiotic forms in large 3D electron microscopy images. This segmentation is required in order to reconstruct in 3D and quantify the morphometrics of key organelles, such as the chloroplast. In particular, the complexity of the cells and size of the stacks render any complete manual annotation extremely time-consuming.
Therefore, obtaining ground-truth annotations by manual annotation for training a deep-learning model is very time-consuming and extremely expensive. New methods for rapid automated segmentation are necessary to unveil the cellular architecture of microalgae.
The solution
In the absence of ground truth for training a deep-learning algorithm, we decided to tackle this challenge using a different approach. We developed a napari plugin to train a Random Forest model on the embeddings of the Segment Anything Model (SAM), guided by a few scribble labels provided by the user. The use of a Random Forest algorithm allows semantic segmentation of multiple types of organelles across the whole stack, with little manual effort.
PROJECT OC1-11
SGEF, a RhoG-specific GEF, regulates lumen formation and collective cell migration in 3D epithelial cysts
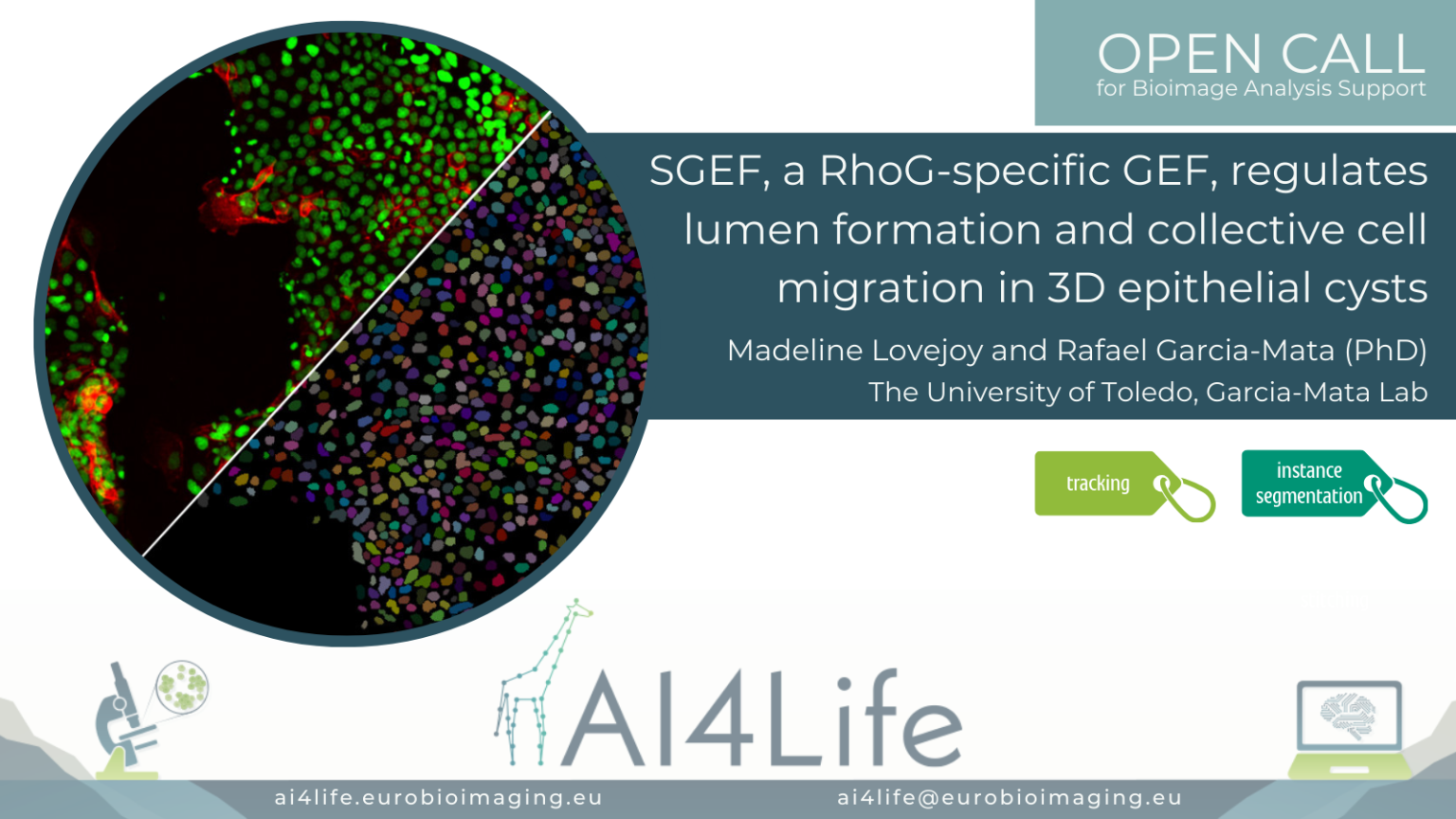
The problem
Researchers from the University of Toledo (USA) are imaging a confluent 2D monolayer of epithelial cells. The monolayer is scratched with a pipette tip, and the video shows the migration of the cells to close the inflicted wound. They would like to compare the behavior of different cell lineages automatically, including cell morphology, for which segmentation and tracking over time of each cell and nucleus is necessary.
The solution
PROJECT OC1-03
Treat Chronic Kidney Disease

The problem
In this project, the researchers, working for RD Néphrologie in Montpellier (France), are studying the effect of Chronic Kidney Disease (CKD) on the density of collagen in specific tissues, such as the heart and kidney. Using mouse and rat models, they extract tissue slices and detect collagen using a biochemical marker.
The solution
In order to estimate the density of collagen in the tissue, we used Labkit to classify each pixel into one of four classes: background, cells, tissue or collagen. Labkit is a Fiji plugin with an intuitive interface that allows labelling pixels in the images and training a random forest classifier. We designed a collection of Fiji scripts to normalise the images, exclude parts of the images from the analysis using masks and perform the quantification. We proposed several ways to obtain the masks for region exclusion, from creating regions of interest in Fiji to using advanced deep learning algorithms such as the Segment Anything Model (SAM).
PROJECT OC1-10
Image-guided gating strategy for image-enabled cell sorting of phytoplankton
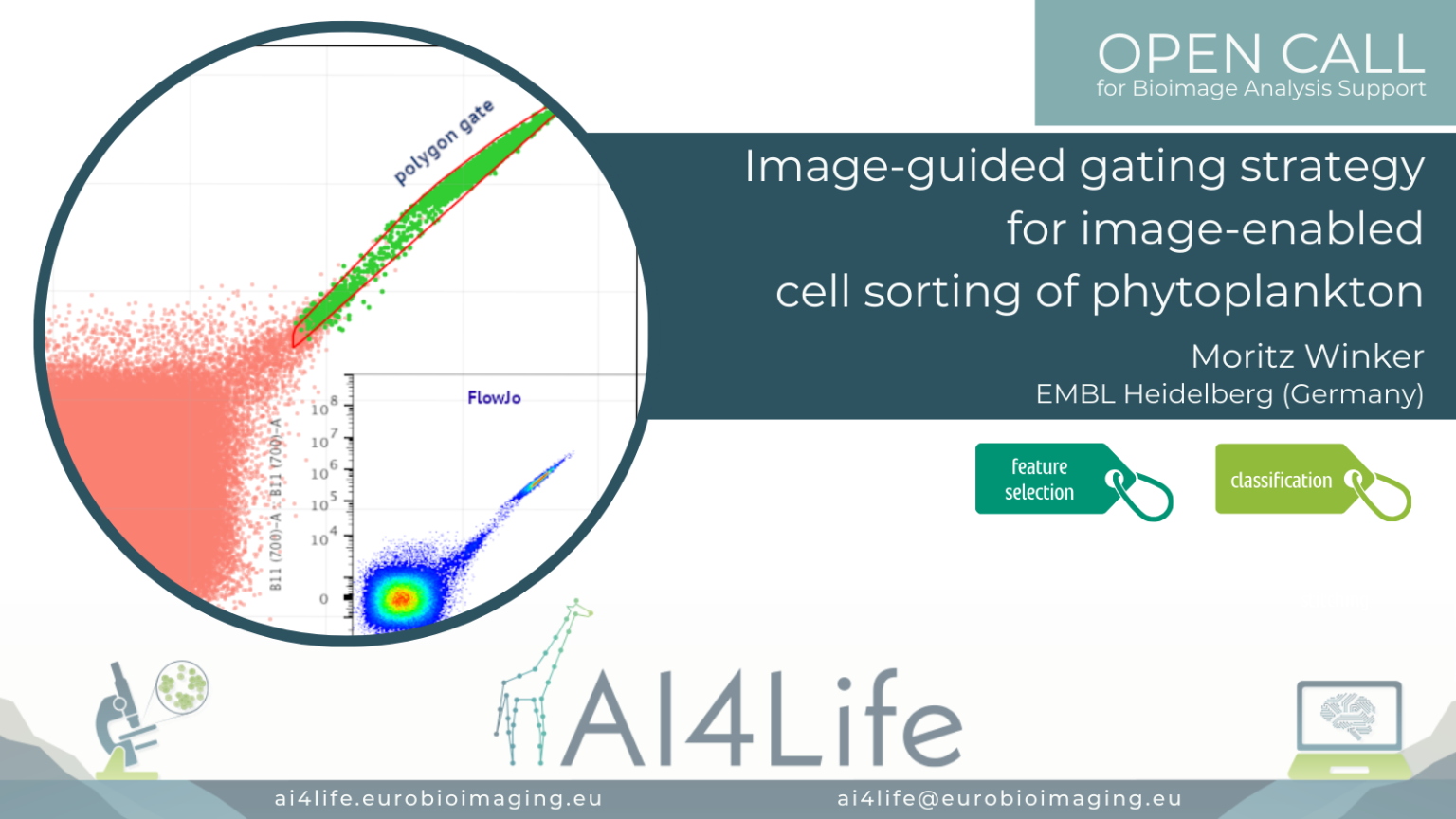
The problem
In this project, a researcher from the European Molecular Biology Laboratory (EMBL) in Heidelberg is using a cutting-edge commercial flow cytometer to sort phytoplankton from lab-grown cultures or field samples. Sorting is traditionally done by selecting features measured by the instrument on the sample and manually drawing a gate defining the range of values in these features that correspond to the cells being selected. However, this new instrument is image-enabled and allows exporting not only traditional features but features derived from fluorescent images as well. In addition, it supports the import of gating strategies to the control software. This opens the door to automated analysis of the features and consequently, the generation of a gating strategy that can be uploaded directly to the flow cytometer.
The solution
To tackle this project, we developed a simple feature selection approach, then generated polygon gates out of 2D histograms while using a similarity threshold between samples. Finally, we explored the upload of gating strategies to the instrument as a proof of principle.
PROJECT OC1-74
Leaf tracker plant species poof

The problem
Researchers from Wageningen University are cultivating various plants in a unique growing facility called NPEC. In each of the NPEC chambers, plants experience identical conditions in terms of light, water, and nutrients. Positioned above the platform, a camera captures images of each plant at specified intervals over several weeks, enabling comprehensive monitoring of their growth and development. This camera system incorporates measurements of RGB data, as well as data from fluorescence, thermal, and hyperspectral cameras. However, the original system and analysis involve averaging measurements from both older and younger leaves of each plant. To gain a deeper understanding of leaf physiology and development under varying light conditions, a quantitative analysis of individual leaves is necessary. Thus, the objective of this project is to develop an AI model capable of analyzing each leaf throughout its developmental stages.
The solution
To crop individual plants, we used PlantCV – a popular library for plant image processing. Then, we segment the individual leaves on the central plant using an instance segmentation deep learning network. Finally, we track the resulting segmentations over time using existing tracking packages written in Python.
PROJECT OC1-47
Identifying senescent cells through fluorescent microscopy

The problem
The solution
Data
Link to BIA
PROJECT OC2-11
Automated species annotation in turbid waters

Project description
The project’s aim was to create an automated image-annotation pipeline to monitor the ecological status of vulnerable marine habitats according to the goals of multiple European Directives. The variability of the image quality and the diversity of the species made this project challenging.
Training a semantic segmentation model was not possible because they did not have enough ground-truth masks. However, they had a good amount of point annotations, which we could use as the prompt for the SAM2 model. We provided a pipeline for applicants to utilise their point prompts to produce instance segmentation masks automatically, and then export the selected masks into COCO format, which was compatible with the BIIGLE that they are using as their main image and annotations platform.
All details about this project, including how to install the required software and conduct the analysis pipeline that we developed in collaboration with the OC Applicants, can be found on GitHub. The FAIR data for this project is accessible via this IDR data record.
PROJECT OC2-13
Optimising calcium image acquisition with machine learning denoising algorithms
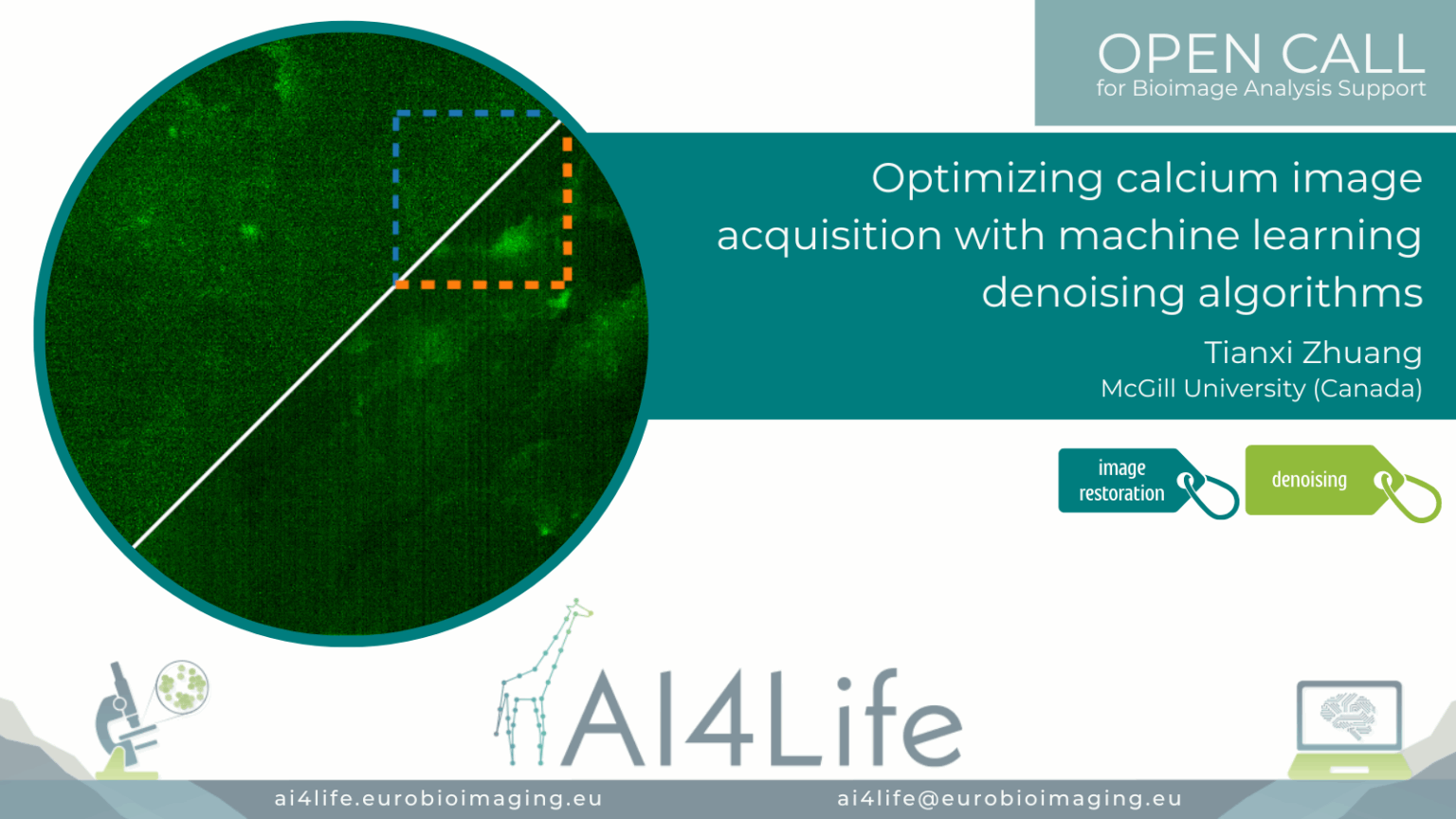
Project description
The project developed a deep-learning–powered pipeline to denoise calcium imaging data acquired from in vivo brain recordings. Leveraging U-Net–based models and temporal-aware filtering, the pipeline enhances weak calcium signals in fast imaging sessions while preserving true biological activity patterns. Integrated with Napari for visual QC and implemented in Jupyter notebooks, the method improves signal-to-noise ratio, aiding downstream spike inference and activity quantification.
All details about this project, including how to install the required software and conduct the analysis pipeline that we developed in collaboration with the OC Applicants, can be found on GitHub. The FAIR data for this project is accessible via this Zenodo data record.
PROJECT OC2-14
Diatom symbioses at planetary scale

Project description
The project developed a scalable image analysis pipeline for quantifying diatom-bacteria symbiotic interactions using large-scale plankton imaging datasets. Using a combination of classical segmentation (watershed, thresholding) and machine learning (pixel classifiers), the workflow identifies diatom frustules and co-localised symbiotic bacteria across diverse oceanic sampling stations. Geographic metadata is integrated to map symbiosis patterns globally. Outputs include co-occurrence statistics and spatial distribution overlays.
All details about this project, including how to install the required software and conduct the analysis pipeline that we developed in collaboration with the OC Applicants, can be found on GitHub.
PROJECT OC2-21
Automated segmentation of actin filaments in intact cells (EM)
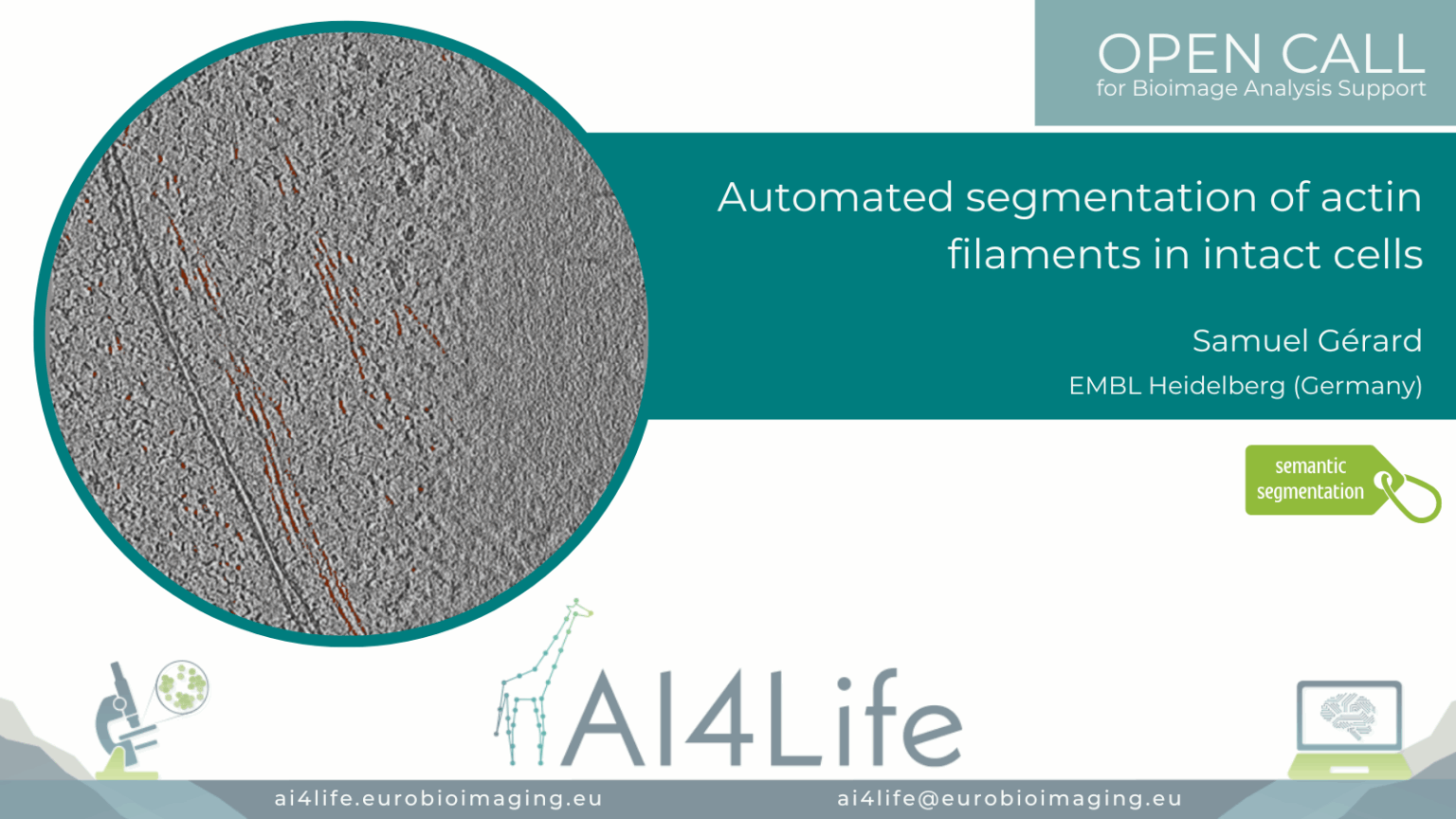
Project description
The project developed an electron microscopy image analysis pipeline focused on dense filamentous actin (F-actin) segmentation in whole-cell tomograms. It integrates pre-trained deep neural networks fine-tuned with human-annotated micrographs and applies post-processing to resolve filament bundling and branching points. The pipeline generates 3D reconstructions of cytoskeletal organisation, aiding studies on actin-mediated cell mechanics. Outputs include annotated segmentations, filament orientation metrics, and filament length distributions.
All details about this project, including how to install the required software and conduct the analysis pipeline we developed in collaboration with the OC Applicants, can be found on GitHub.
PROJECT OC2-23
Enhancing the effective resolution of intravital microscopy with digital expansion microscopy
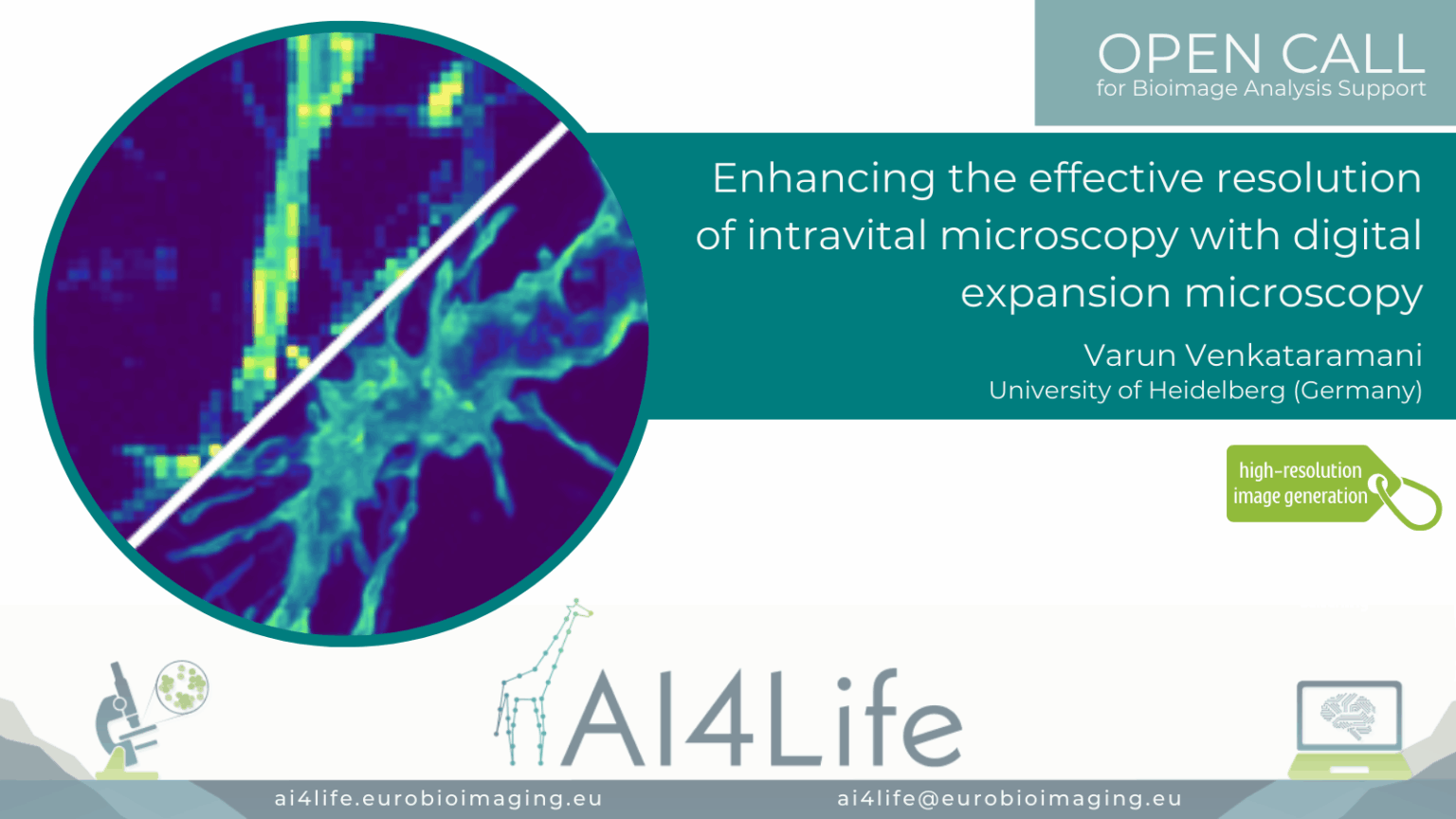
Project description
The project developed a deep learning pipeline to enhance in vivo two-photon microscopy images to super-resolved quality equivalent to 4× physical expansion microscopy, overcoming diffraction limits without the need for physical sample enlargement. By creating a synthetic training dataset from expansion microscopy images paired with simulated low-resolution counterparts, the model learned to recover fine subcellular features such as tumour microtubules and membrane protrusions in glioblastoma xenograft models expressing membrane-targeted GFP. Importantly, the pipeline integrates uncertainty estimation methods like variational autoencoders, supporting more reliable biological interpretation and downstream analysis.
All details about this project, including how to install the required software and conduct the analysis pipeline that we developed in collaboration with the OC Applicants, can be found on GitHub. The FAIR data for this project is accessible via this IDR data record.
PROJECT OC2-26
Ultrastructural protein mapping through Correlation of Light and Electron Microscopy
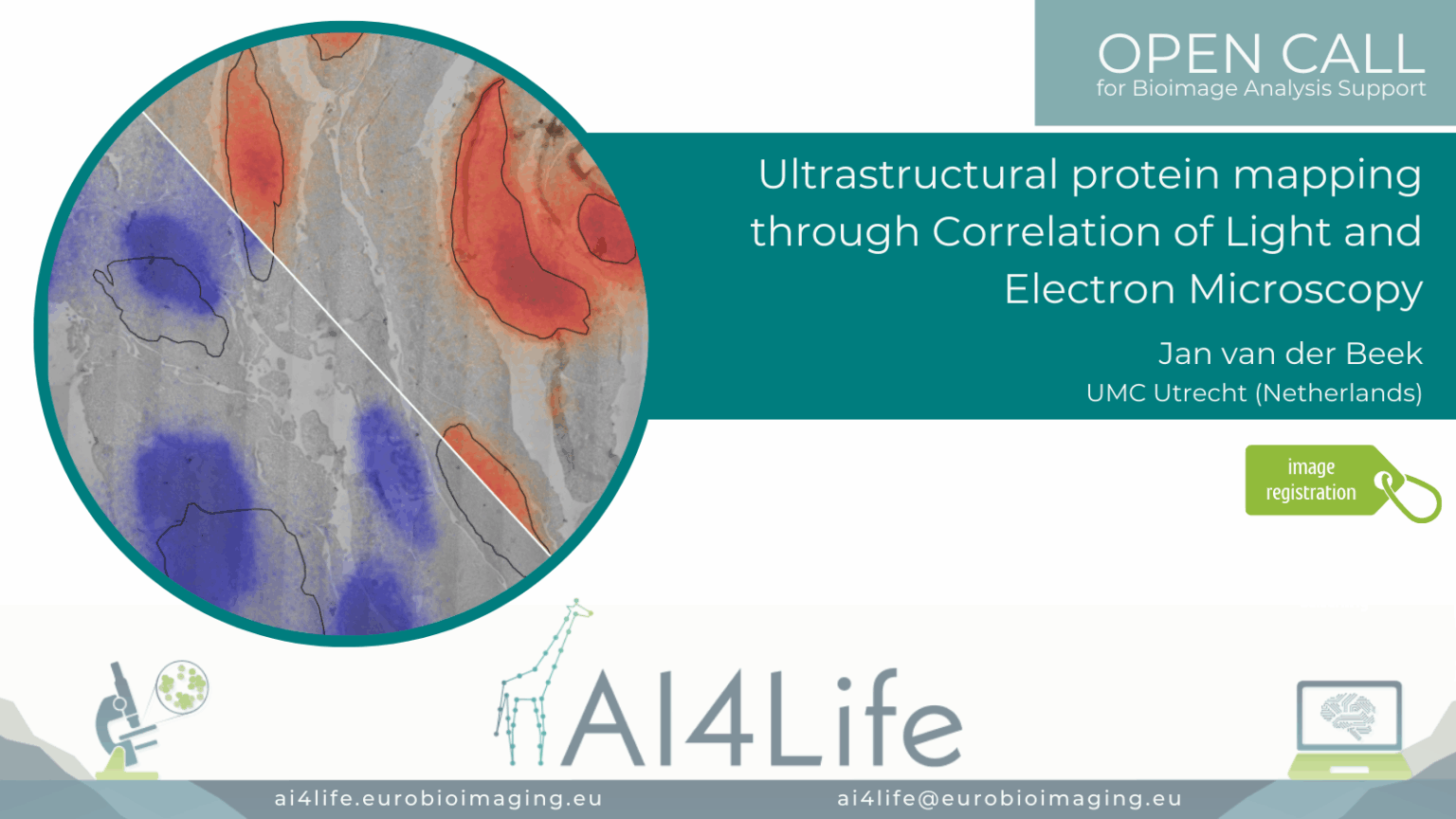
Project description
The project developed a correlative light and electron microscopy (CLEM) workflow for ultrastructural mapping of protein distributions in cells. It aligns high-resolution TEM images with fluorescence microscopy channels using fiducial markers, then overlays segmented protein signals onto electron-dense structures. Registration is automated through landmark-based matching in the Napari plugins. Quantitative readouts include protein localisation maps, proximity to organelles, and relative fluorescence intensities.
All details about this project, including how to install the required software and conduct the analysis pipeline that we developed in collaboration with the OC Applicants, can be found on GitHub. The FAIR data for this project is accessible via this IDR data record.
PROJECT OC2-38
NeuroScan – A 3D human motor neuron disease platform for high-throughput drug screening
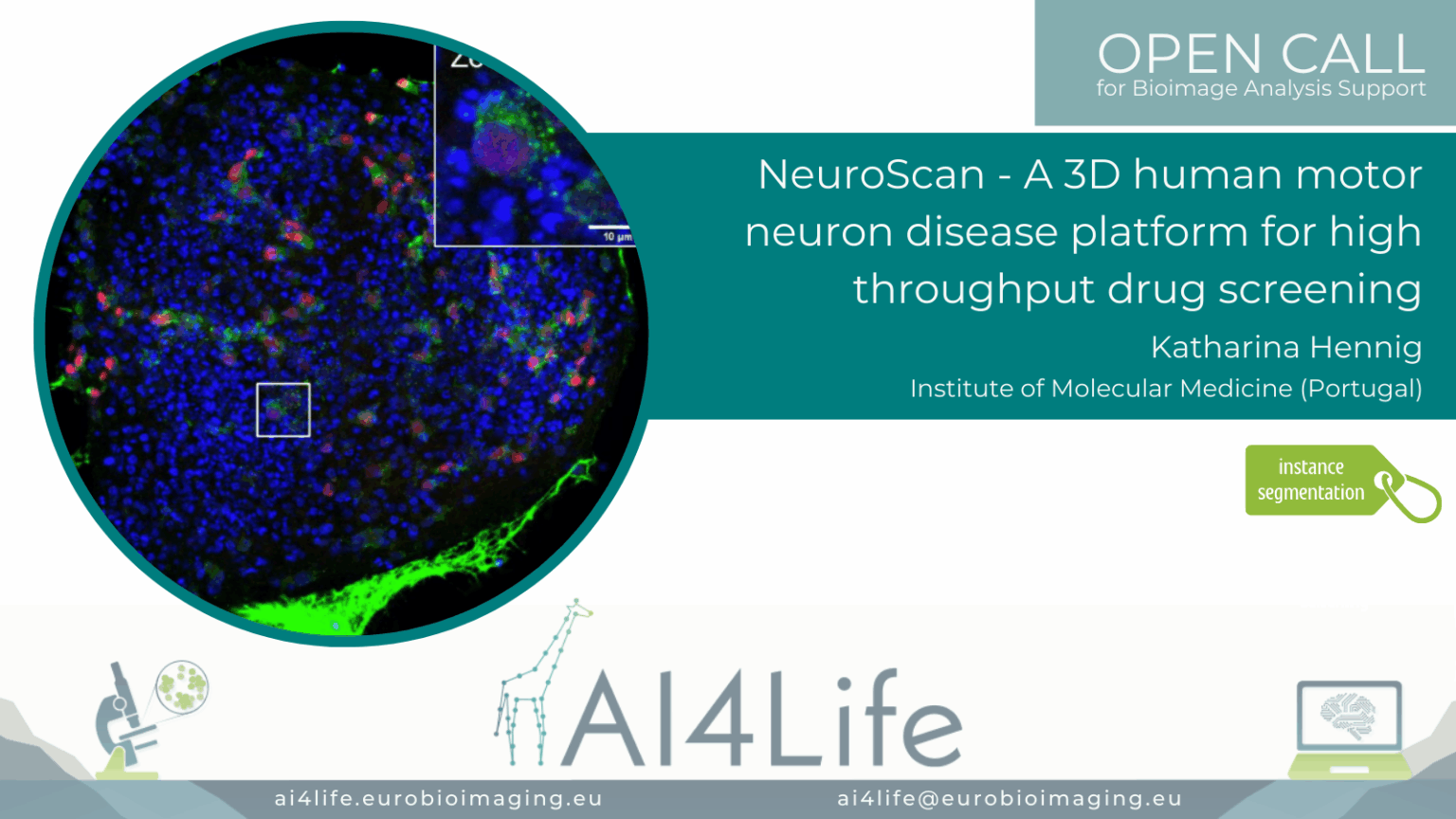
Project description
The project developed a scalable 3D screening platform for analysing human motor neuron disease (MND) models using high-content microscopy. It segments neuron soma and neurite networks in 3D culture systems using hybrid models combining Cellpose and volumetric U-Nets. The workflow quantifies neurite outgrowth, soma size, and disease markers across drug-treated samples. Outputs support phenotype-based screening and therapeutic profiling in patient-derived iPSC models.
All details about this project, including how to install the required software and conduct the analysis pipeline that we developed in collaboration with the OC Applicants, can be found on GitHub. The FAIR data for this project is accessible via this IDR data record.
PROJECT OC3-03
Improving nuclei segmentation using Cellprofiler and StarDist
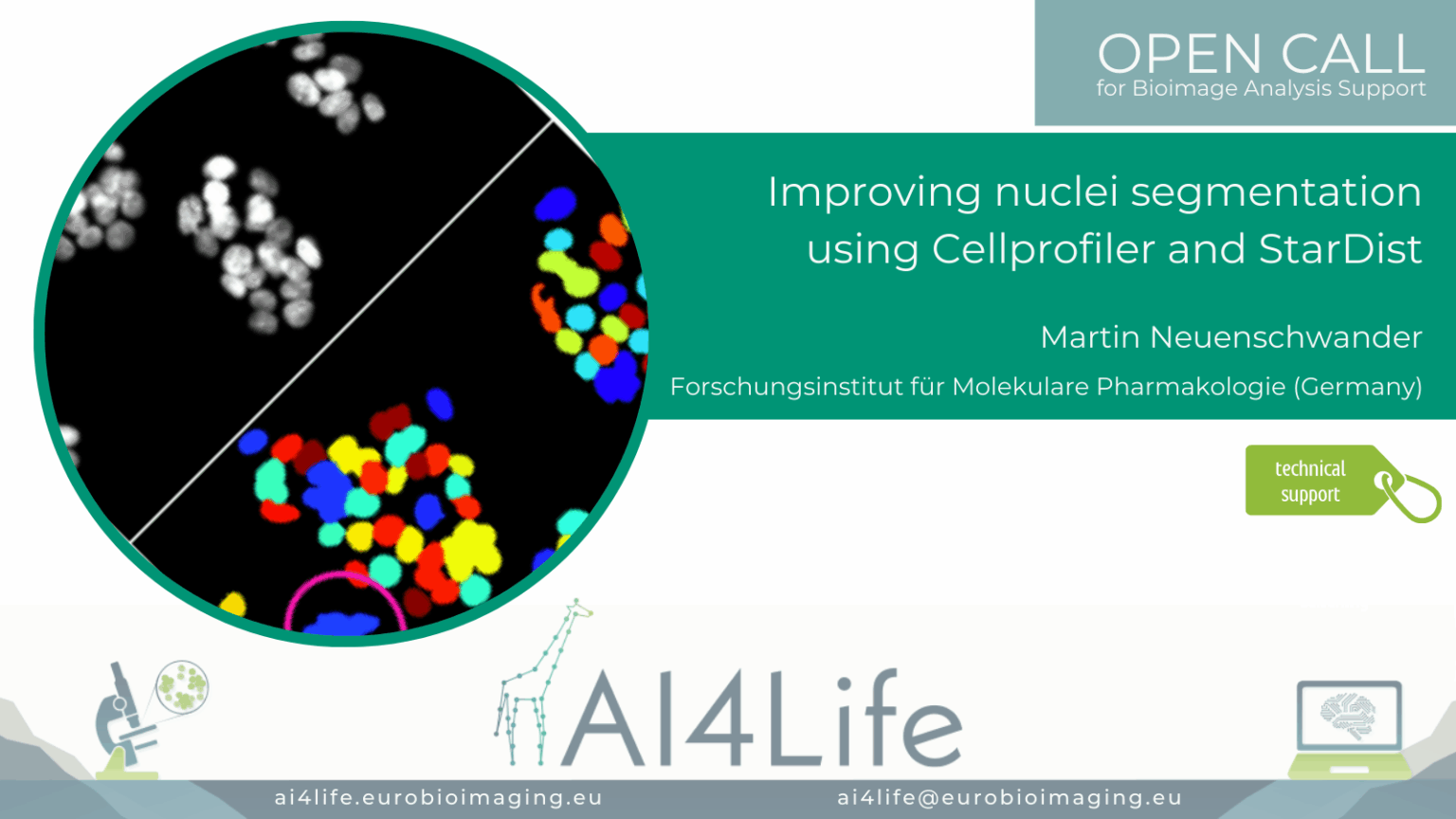
Project description
A group of researchers in Germany is using the Cell Painting assay to image different cellular structures with multiplexed dyes to understand which of them are affected by the compound. The automated image acquisition and computational feature extraction generate rich, quantitative fingerprints of each compound’s effect on cell morphology and subcellular organization.
The objective is to create an open database of the compounds with their phenotypic profile, also publishing the acquired images in the Bioimage Archive. This comprehensive dataset has the potential to help researchers across the world cluster compounds by mode of action, predict biological targets, and discover novel drug leads.
The automatic image-analysis workflow is implemented in CellProfiler. Because HepG2 cells are small and often form dense clusters, accurate segmentation can be challenging and typically requires specialized deep learning models, such as StarDist, to reliably delineate individual cells. To integrate StarDist and other BioImage.io – compatible networks into our pipeline, we have enhanced deepImageJ to run directly within CellProfiler, ensuring seamless execution of advanced segmentation and feature-extraction routines.
All details about this project, including how to install the required software and conduct the analysis pipeline that we developed in collaboration with the OC Applicants, can be found on GitHub.
PROJECT OC3-11
Matrix Motility Map (3DM³)
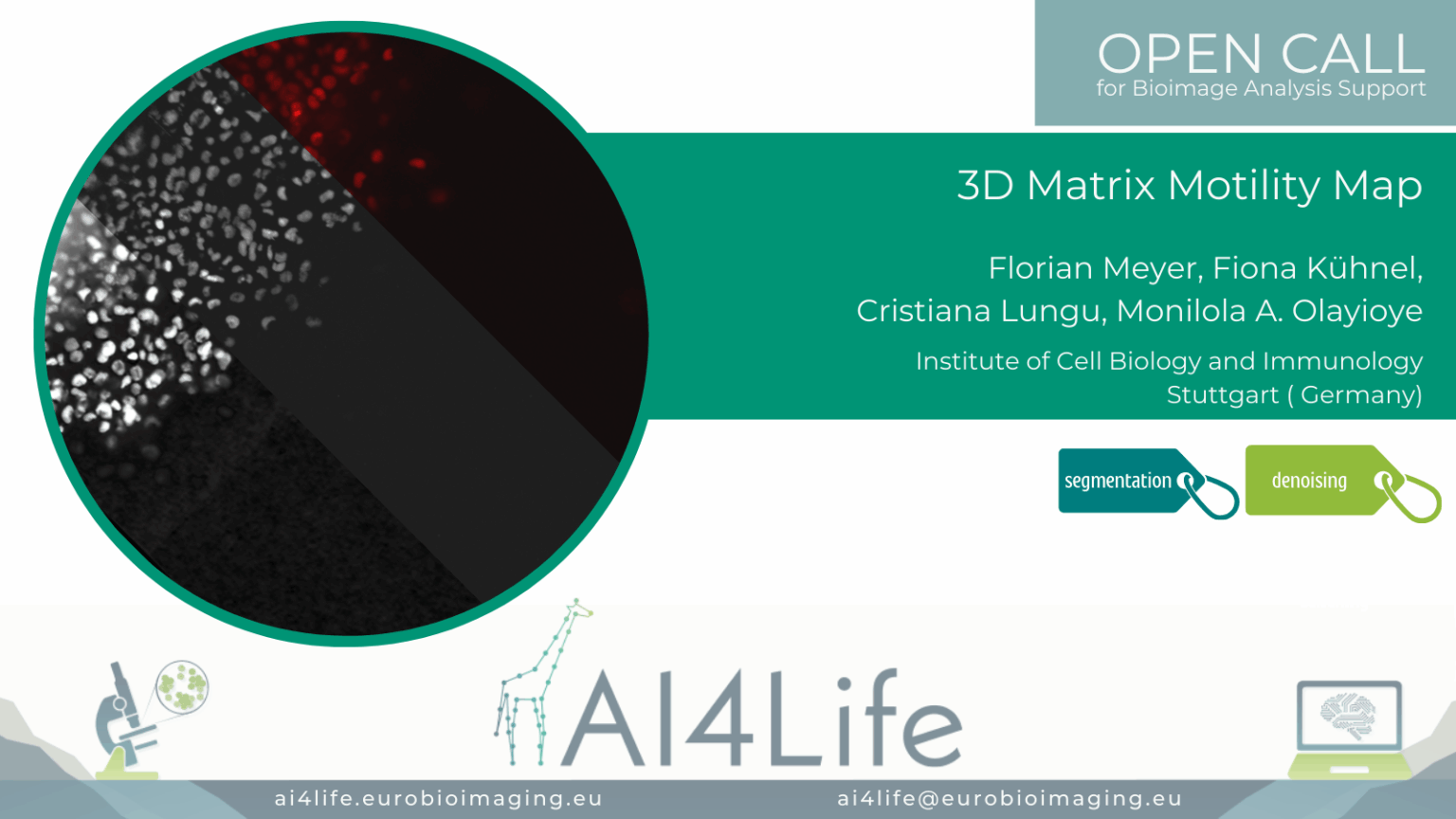
Project description
This project focuses on live-cell imaging of breast tumour spheroids embedded in collagen to analyze cancer cell migration, shape, and interaction with ECM architecture. This project aims to analyze cancer cell dissemination in 3D matrices, which is driven by interactions between cells and the extracellular matrix (ECM), particularly collagen.
The dataset includes 5D live imaging (X, Y, Z, T, C) acquired using widefield and confocal microscopy.
All details about this project, including how to install the required software and conduct the analysis pipeline that we developed in collaboration with the OC Applicants, can be found on GitHub.
PROJECT OC3-14
The speed of life in trees

Project description
The goal of this study was to automatically measure the fractional areas of vessels, axial and ray parenchyma, and fibers across the entire image, utilising a semantic segmentation pipeline.
Handling huge whole-slide images for 51 tree species and a lack of ground-truth masks for three (of four) cell types made this project challenging.
As a solution, we utilized FeatureForest (Seifi et al (2025)), a tool that was born and developed for a project “Atlas of Symbiotic partnerships in plankton” (project ID 52) of the first open call. With FeatureForest, we could train a model to generate very accurate masks for every cell. Later, the user will be able to assign labels to masks based on cell type. For the vessel cell type, for which we had ground-truth, we trained a deep-learning model combining several species data to be able to predict vessel masks across species.
All details about this project, including how to install the required software and conduct the analysis pipeline that we developed in collaboration with the OC Applicants, can be found on GitHub.
PROJECT OC3-17
Imprints of Wind Disturbances on Wood Anatomy

Project description
Disturbances play a crucial role in forest dynamics. The goal of this project was to analyse differences in wood quality among trees experiencing varying degrees of release (from none to the most intense). The desired outcome was the values of cell lumen area and cell wall thickness for each analysed cell (thousands of cells).
All details about this project, including how to install the required software and conduct the analysis pipeline that we developed in collaboration with the OC Applicants, can be found on GitHub.
PROJECT OC3-18
Detection of Nuclear Pore Complexes with shape variability imaged by DNA PAINT
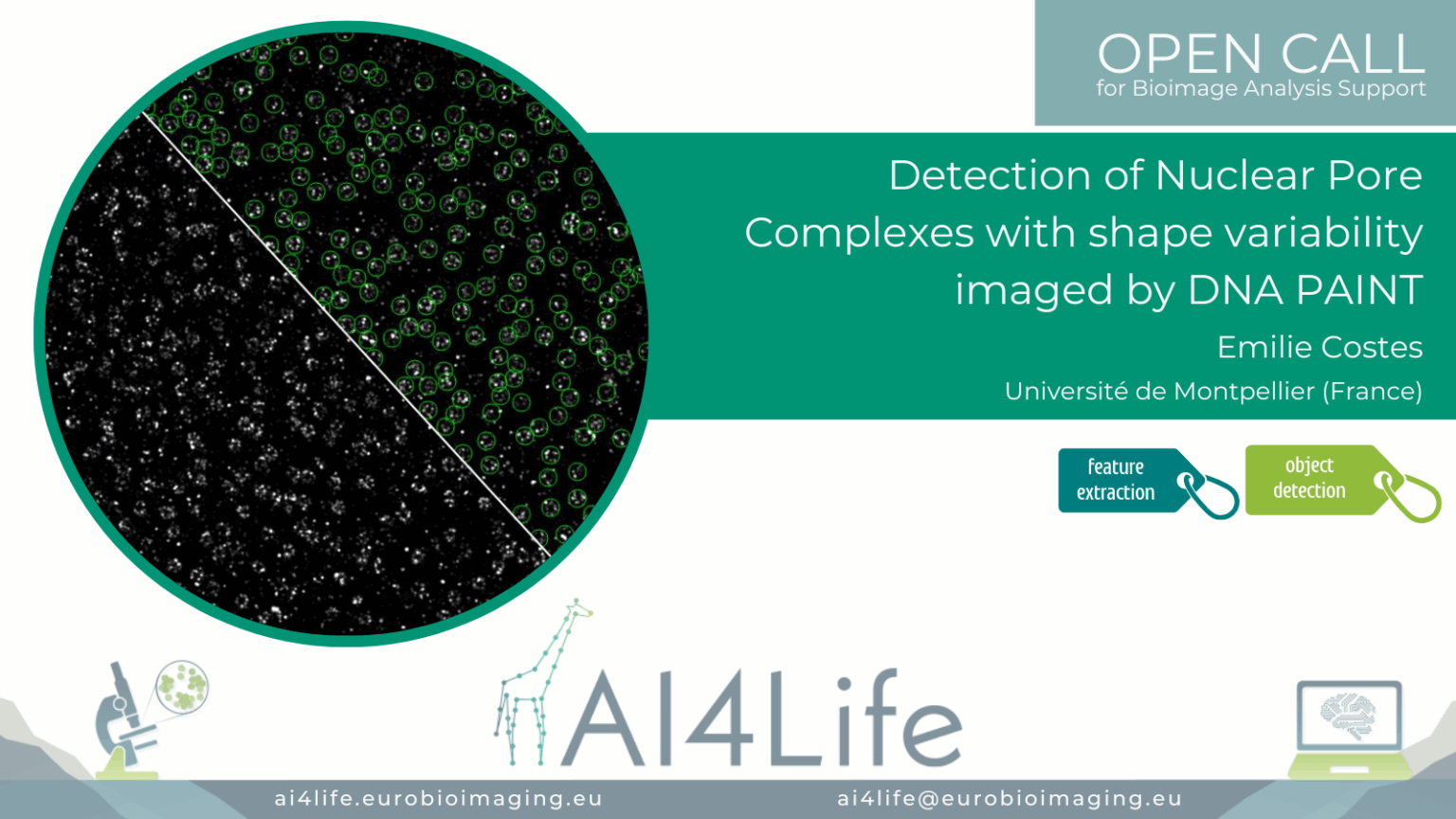
Project description
The project’s aim was to identify nuclear pore complexes in DNA-PAINT images, including those that deviate from the canonical structure in terms of diameter, stoichiometry, and circularity.
All details about this project, including how to install the required software and conduct the analysis pipeline that we developed in collaboration with the OC Applicants, can be found on GitHub.
PROJECT OC3-19
Segmentation of sparse bacteria in human tissue
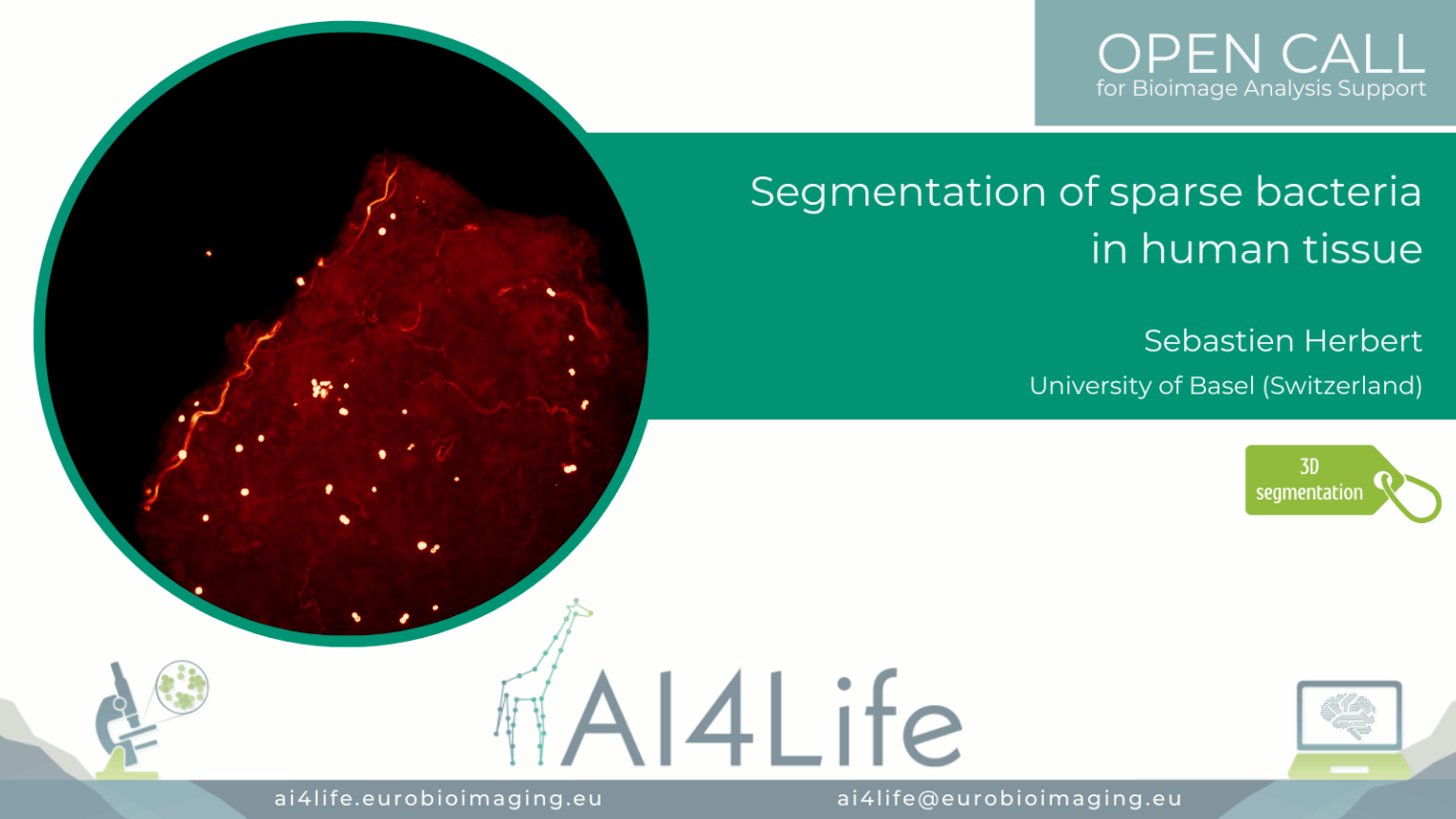
Project description
The project’s goal was to segment and analyse bacteria clusters in human biopsies and characterise the biological microenvironment of niches (intra/extracellular, cell type, cluster size).
The tissue samples are very large (~2 mm² × 60μm), bacteria are sparse, and hollow spheres approximately 1μm in diameter; this makes the instance segmentation of bacteria a challenging task.
The provided solution includes training a 3D StarDist model to process 3D microscopy images of tissue.
All details about this project, including how to install the required software and conduct the analysis pipeline that we developed in collaboration with the OC Applicants, can be found on the AI4Life Website and GitHub.
PROJECT OC3-26
Automatic microtubule doublet picking in tomograms
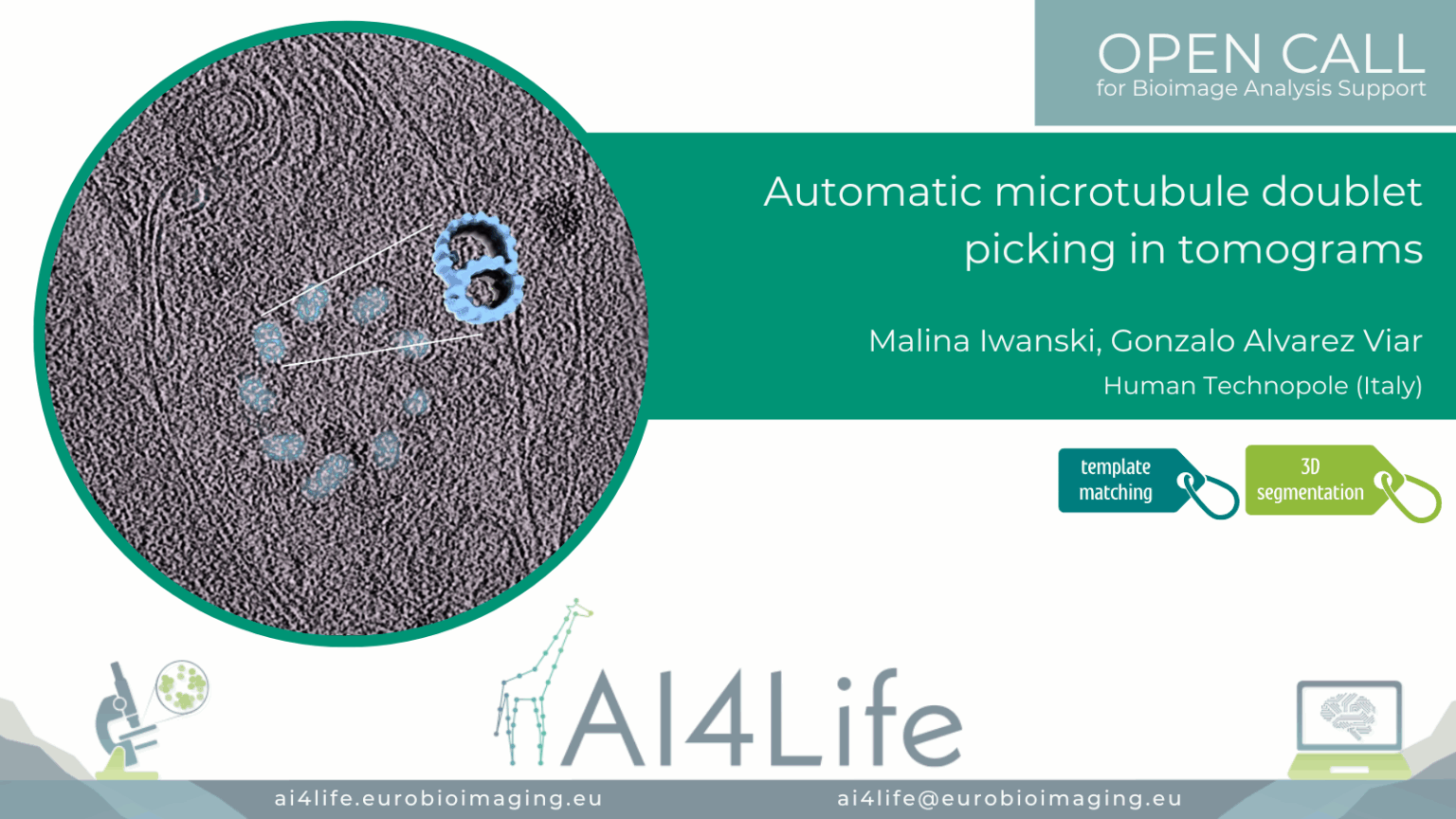
Project description
The project’s aim was to automatically detect and pick doublets with the correct polarity (i.e., all pointing the same) in Cryo-EM tomograms of cilia. Besides the full manual work, another challenge was that the existing software failed to assign polarity correctly.
The raw tilt series was gain and motion corrected using the Warp package. Tomogram alignment and reconstruction were performed using the EMAN2 package. All tomograms were reconstructed using the weighted back projection method and binned by four. For particle picking, the path of each doublet in the bin4 tomograms was modeled in EMAN2. Each doublet was traced by a separate contour made up of ~5 points, which were placed in the center of the filament using the different cross-views of the tomogram. Particles were extracted from the models with a box size enough to encompass the 8 nm repetitive structure present in the filaments and with an overlap between particles of 80%. In order to increase the particle count. Using the orientations traced by the model and the particles extracted, an initial 3D density map of the doublets was generated by averaging all the particles.
All details about this project, including how to install the required software and conduct the analysis pipeline that we developed in collaboration with the OC Applicants, can be found on GitHub.


















