Outcomes of the Second AI4Life Open Call
by Beatriz Serrano-Solano
AI4Life launched its second Open Call on January 22nd, and applications were accepted until March 8th. We’re very happy to share that we received a total of 51 applications for the second AI4Life Open Call!
The applications span a wide range of fields, including developmental and marine biology, as well as cancer research. But that’s not all—we also received submissions from areas such as plant biology, parasitology, ecology, biophysics, microbiology, immunology, and many more.

Why did scientists apply? What challenges are they facing?
Similar to the first open call, most applicants focused on improving their image analysis workflows. However, unlike the first Open Call, we did not offer consultancy as an option to choose, as it is now an implicit step in the process.
Before finalizing project selections, we will hold consultancy calls with a number of project applicants to offer quick guidance that could help researchers find solutions. Projects requiring deep learning support will then be prioritized for the final selection.
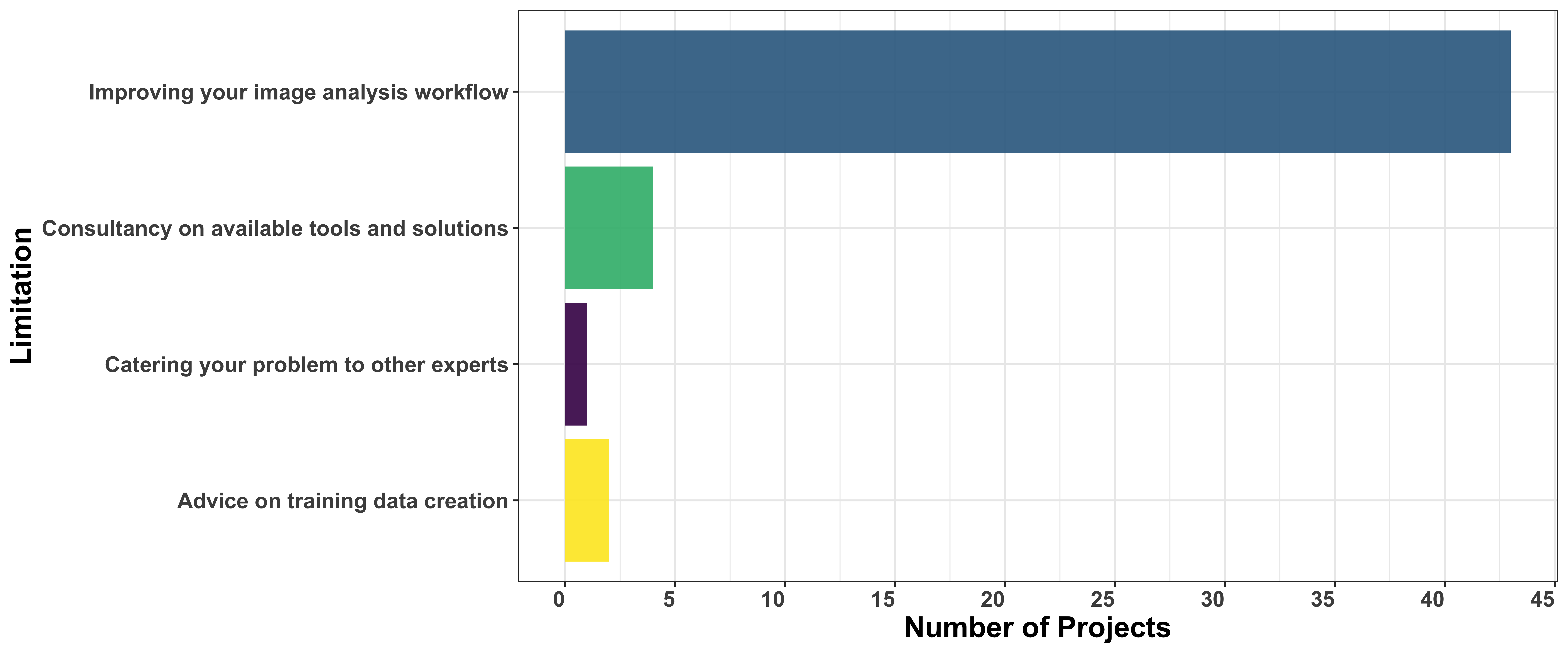
How have applicants addressed their analysis problem so far?
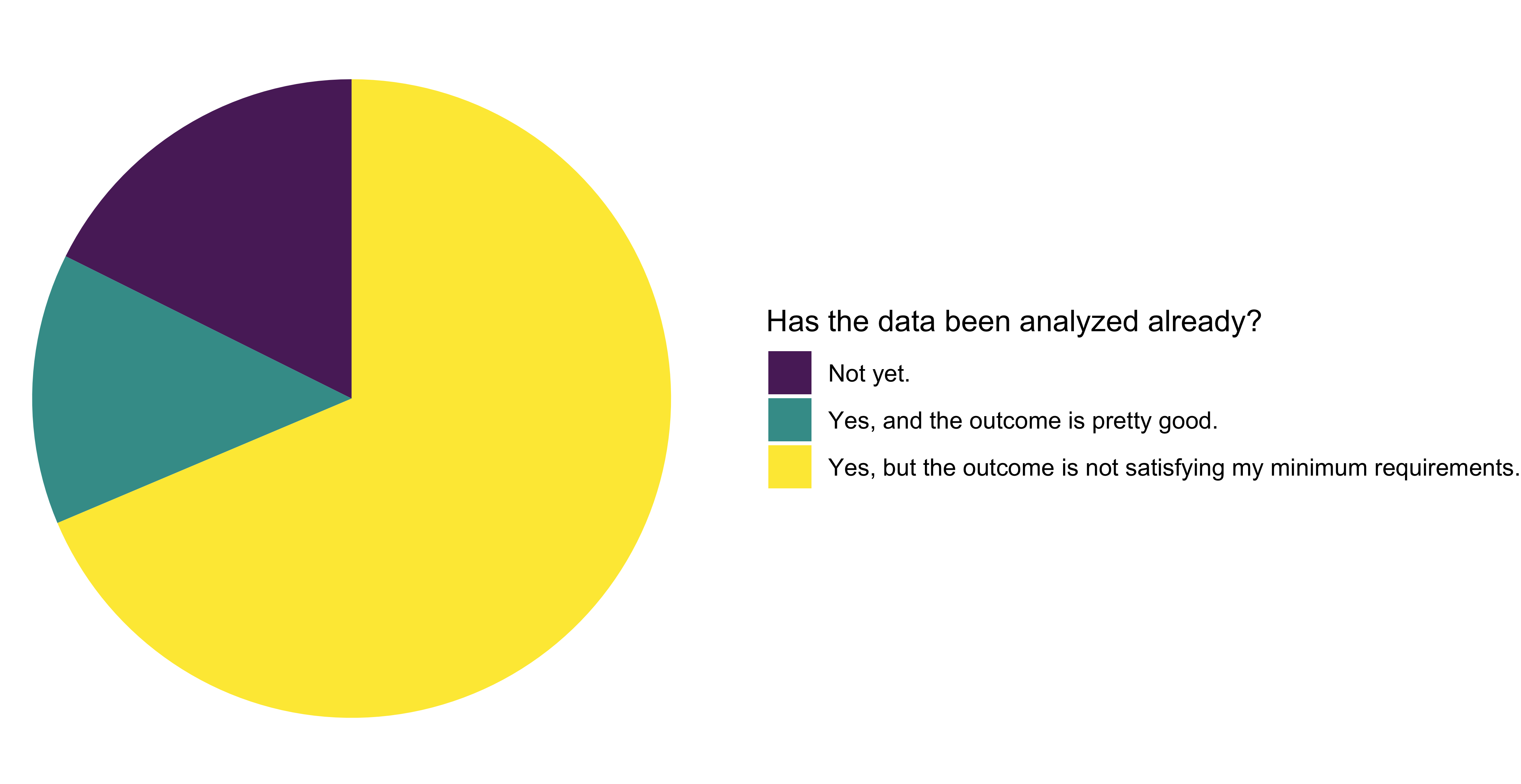
Nearly three-quarters of the applicants had already analysed the data, a higher percentage compared to last year. However, only one-fifth of them had not done so yet. The remaining applicants had already analyzed the data and found the outcome satisfactory. Last year, twice the fraction of projects were satisfied with the outcome compared to this time.
When asked about the tools used to analyze their image data, Fiji and ImageJ remain the most popular choices, followed by custom scripts and commercial software.

What kind of data and what format do applicants deal with?
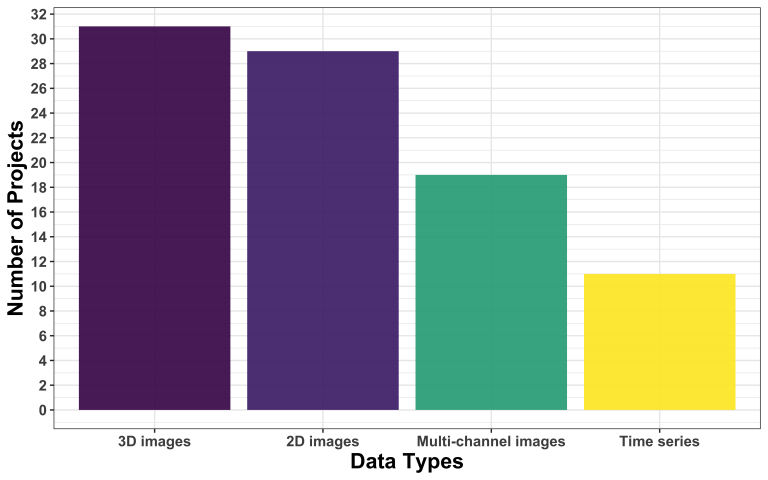
In terms of data format, we observe a change in the trend compared to the first call. Now, 3D images are slightly more frequent than 2D images, reversing the trend we observed last time.
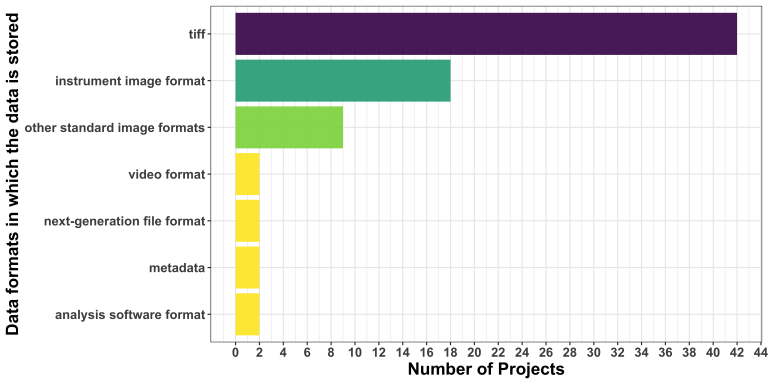
TIFF remains the most popular format, while the second group comprises more proprietary file formats. We are thrilled to observe that the Next Generation File Format (NGFF) has appeared this time!
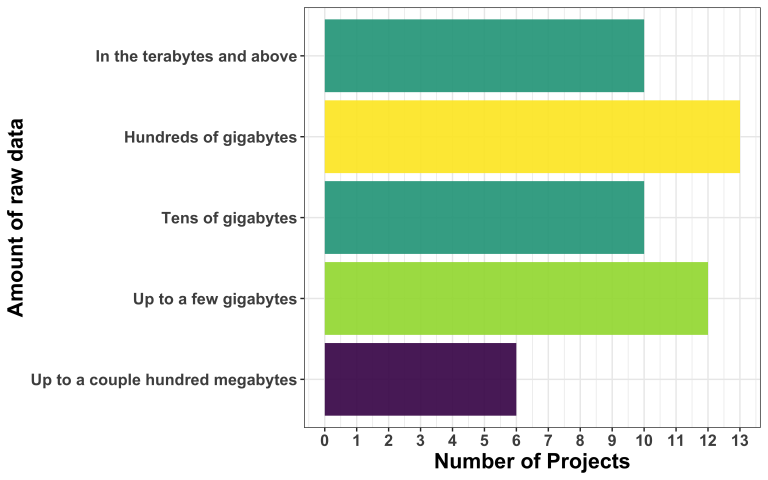
How much project-relevant data do the applicants bring?
The majority of projects (45 out of 51) manage data in the order of gigabytes or above. Only 6 projects involve data sizes up to a few hundred megabytes. This highlights the prevalence of larger-scale datasets within the applicants, suggesting a growing demand for proper data management and processing capabilities.
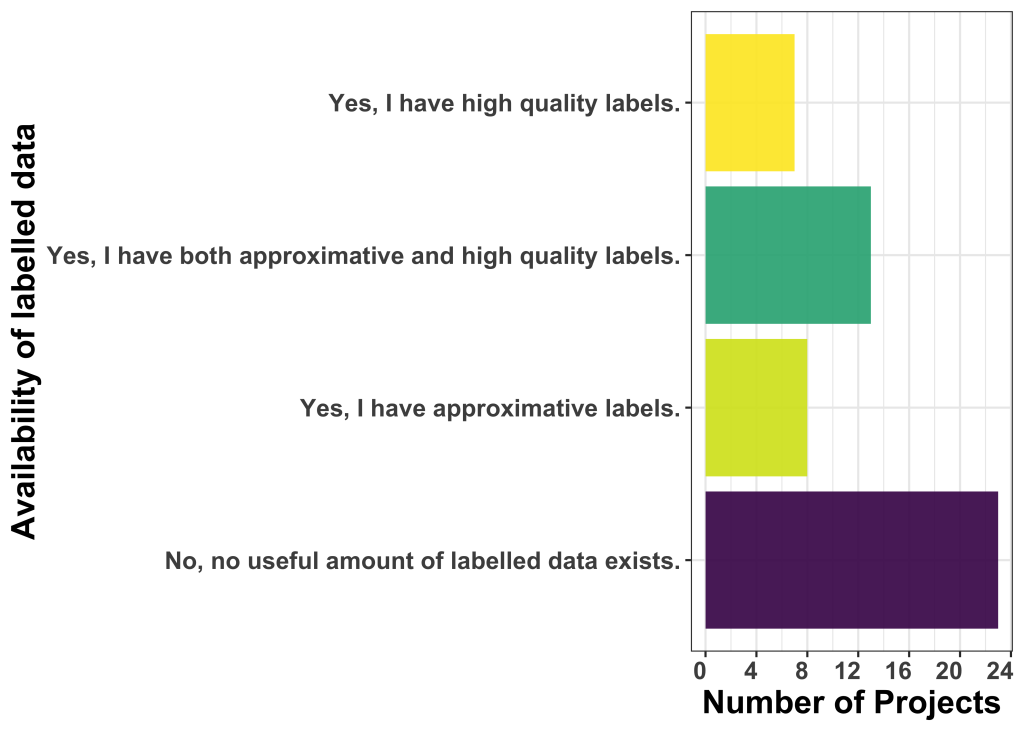
Is ground truth readily available?
When asked about the availability and percentage of labelled data, approximately half of the applicants (23 out of 51) reported a lack of sufficient labelled data. Despite a variance in the definition of labelled data compared to the first open call, we observe a comparable trend. Additionally, the proportion of projects indicating high-quality labels has decreased compared to the previous call.
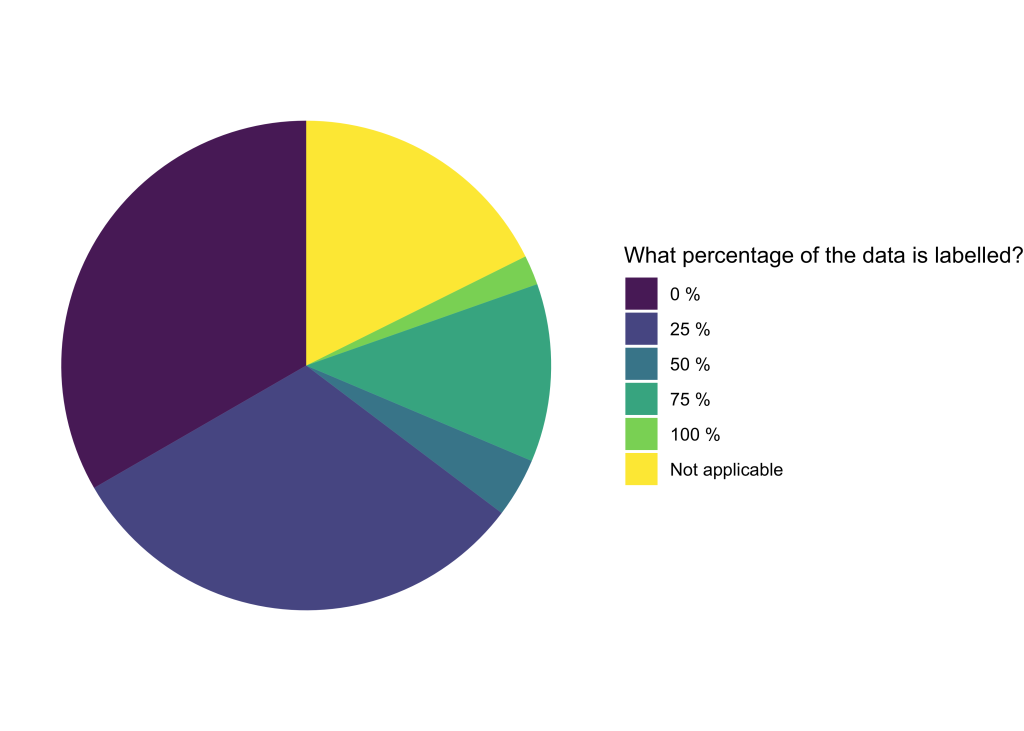
In the application form for this second open call, we introduced an additional question regarding the percentage of labelled data. Over three-quarters of the projects have less than 25% of their data labelled, while approximately 20% of the projects have more than half of their data labelled.
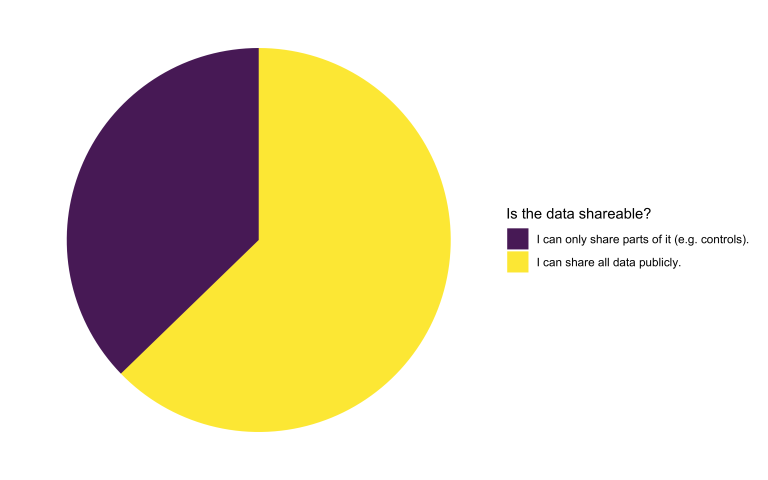
Is the available data openly shareable?
Interestingly, the ratio of projects that can provide access to all their data remains consistent compared to the first open call. This time, we’ve introduced the option of exclusively sharing the controls. The reason behind this decision is that projects unable to share any data are not eligible for support in applying deep learning to their research project.
What’s next?
We have completed the eligibility check and expert reviews for the submitted projects. Projects were reviewed by 17 experts, resulting in an average of 3 reviews per project. The reviews have been aggregated, and scores have been computed to rank the projects based on these reviews. As a result, a preselection of projects has been made, and these applicants will soon be notified.
What to expect from the consultation phase?
During the consultation call, selected applicants will have the opportunity to engage with experts who will provide insights, tips, existing tools and recommendations to guide their project. Following the consultation phase, a subset of projects will be chosen to receive expert support based on their potential and need for deep learning support.
