What’s next for AI4Life?
by Dorothea Dörr, Beatriz Serrano-Solano
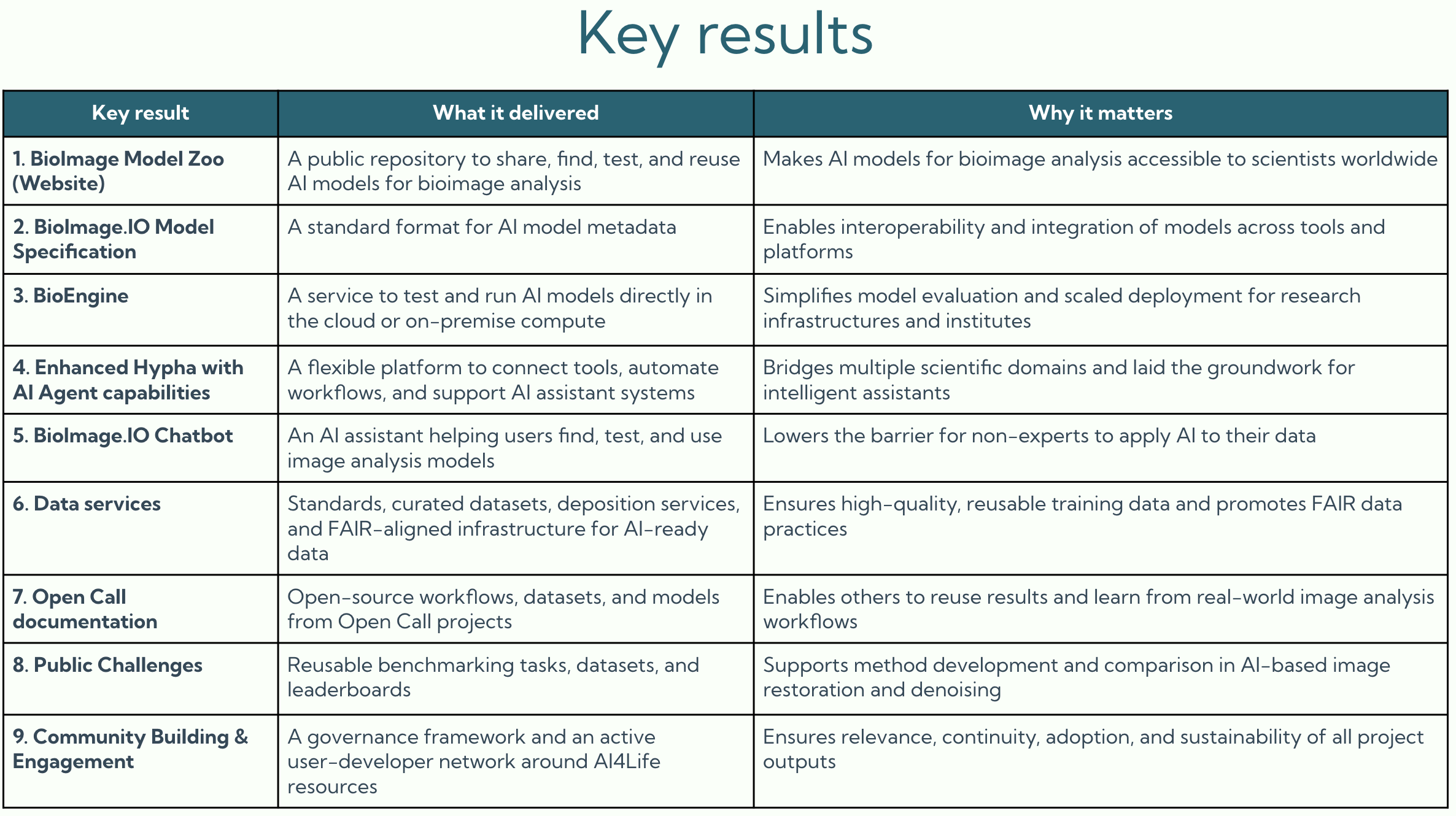
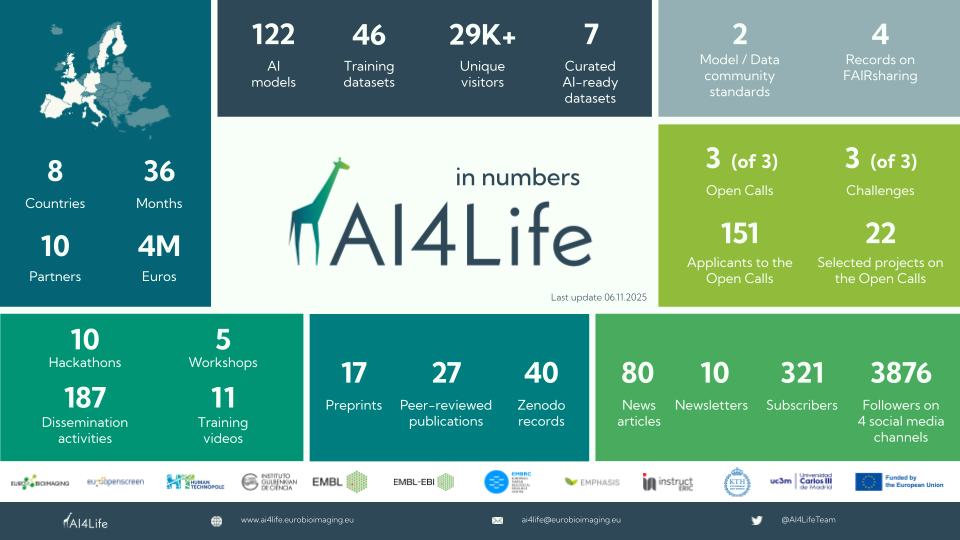
The AI4Life project has officially come to an end, but its impact and activities are far from over. Together with our partners, we’ve built the foundations for an ecosystem that will keep growing and supporting the image analysis community.
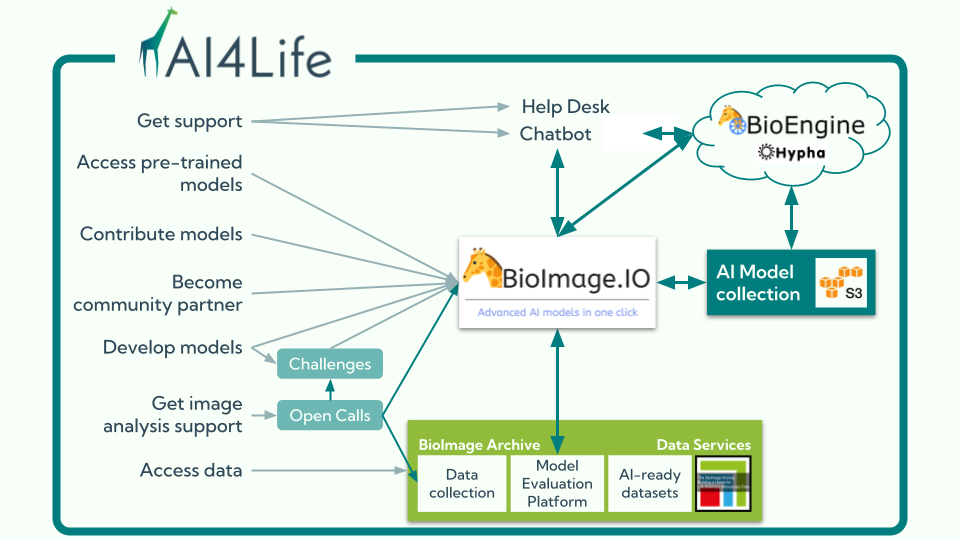
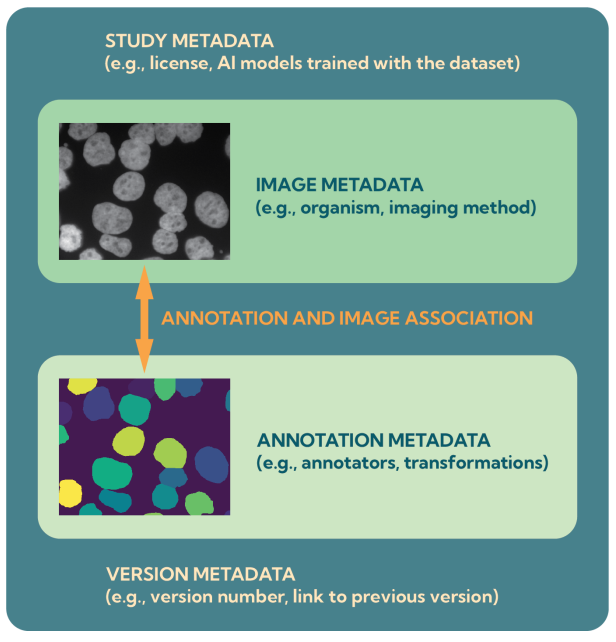
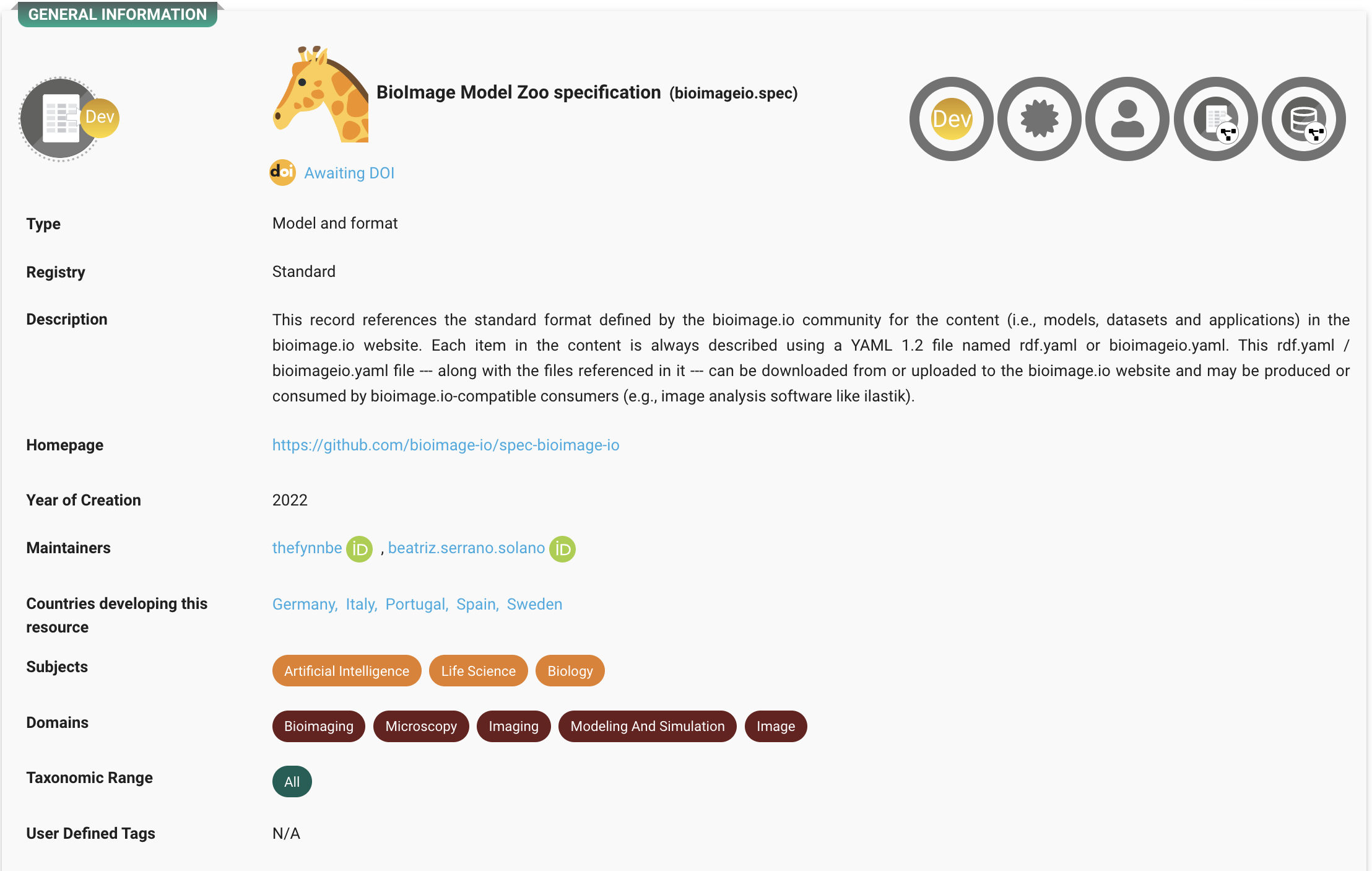
At the heart of this ecosystem is the BioImage Model Zoo (BMZ), a community-driven platform that makes pretrained AI models for bioimaging openly available and easy to use. The BMZ is designed for life scientists, imaging facilities, healthcare researchers, and industry partners who want to apply AI methods to their data.
The ecosystem moving forward
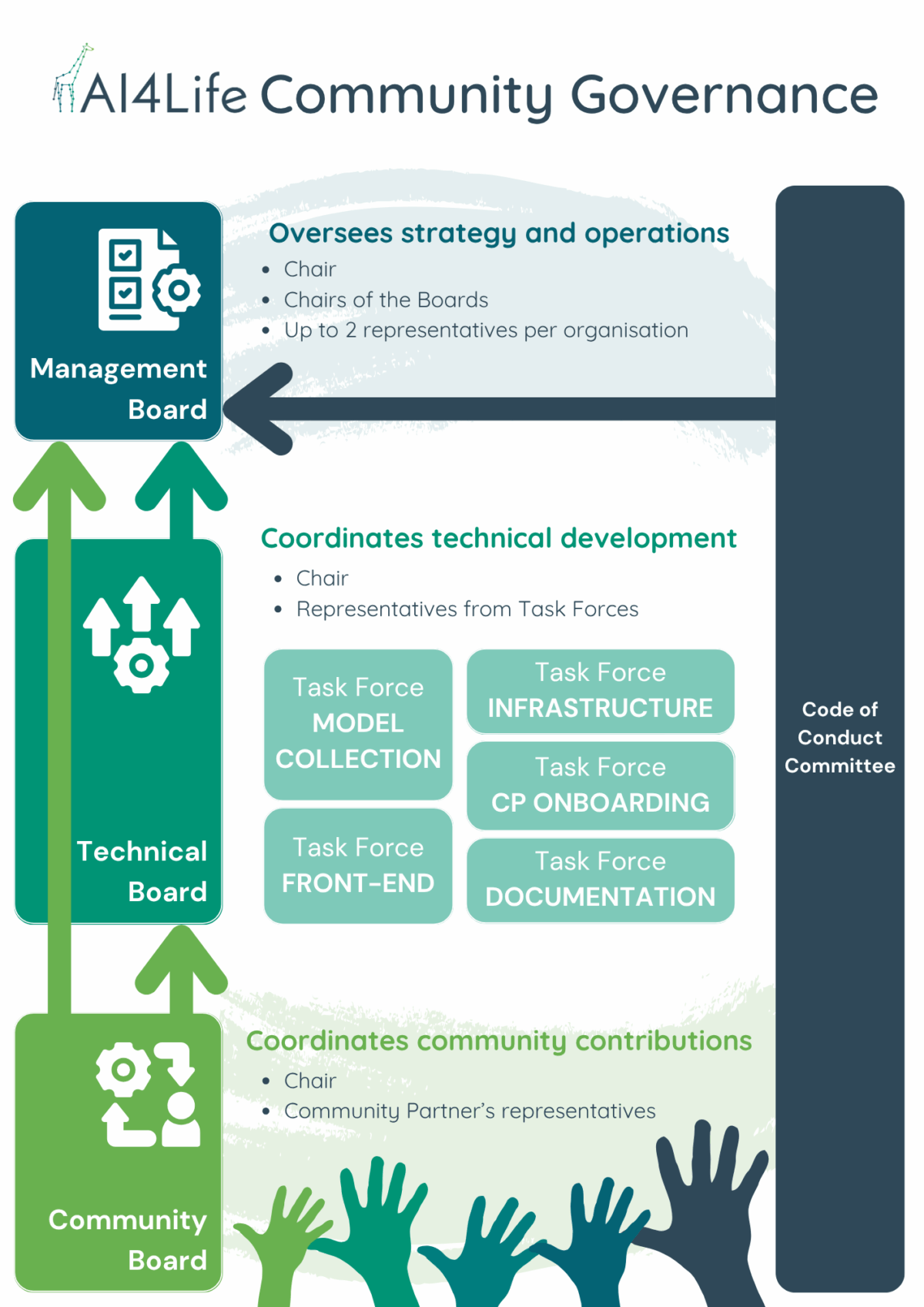
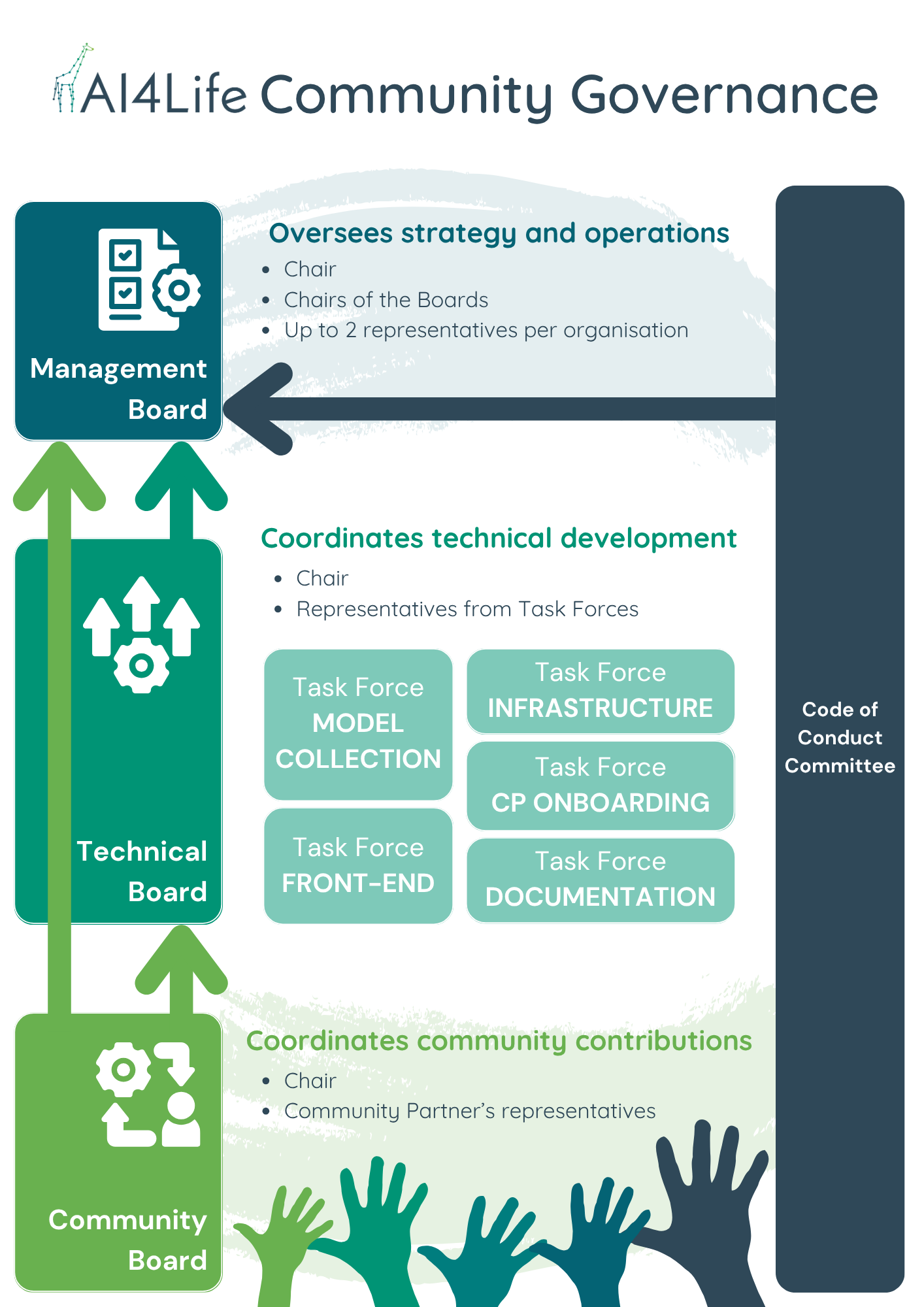
To ensure long-term sustainability, the BMZ is guided by a governance structure coordinated by Euro-BioImaging, with the founding partners KTH/SciLifeLab, EMBL, HT, UC3M, ITQB-NOVA, and EMBL-EBI.
Governance is organised into several boards that reflect different needs:
- Management Board: sets the overall strategy and roadmap.
- Technical Board: drives development and infrastructure.
- Community Board: gathers feedback, coordinates contributions, and represents the broader community.
How you can contribute
The BMZ thrives on community input, and there are many ways to get involved:
- Submit models: share your AI models with the community.
- Collaborate as a partner: institutions can join as Community Partners, gaining a voice in governance and helping steer the future of the platform.
- Join discussions: contribute feedback on features and standards.
- Participate in task forces: help shape technical or community-focused developments.
- Promote and teach: use the BMZ in your courses, workshops, or talks.
By continuing this work, we ensure that the knowledge, tools, and infrastructure built during AI4Life remain available for the scientific community, fostering innovation for years to come.
Stay connected
Community governance is open and transparent. Anyone can join our weekly Wednesday meetings at 16:00 CE(S)T:
- First week of the month: Community Board (with current and prospective Community Partners).
- Other weeks: Technical Board (with Task Forces spun up as needed).
The same Zoom link will be used for all meetings, and agendas are kept in HackMD, and meeting notes are posted to GitHub for wider visibility (details here).
Follow Euro-BioImaging and our partners to hear more about upcoming developments, community opportunities, and how you can get involved in shaping the BioImage Model Zoo.
And don’t forget to subscribe to the Euro-BioImaging Newsletter!