Engaging with AI4Life made easier


by Beatriz Serrano-Solano
We’ve launched a new section on our website dedicated to guiding you on how to engage with our project.
Are you looking to participate, collaborate, or simply learn more about AI4Life? Our new section has all the answers. Find out how you can contribute, connect, and engage with us effortlessly!
Explore the new section: https://ai4life.eurobioimaging.eu/engage/


by Daniel Franco
The Bioimage Analysis software BiaPy has officially joined the BioImage Model Zoo as a Community Partner! This means that the BiaPy software supports the BioImage.io format for deep learning models.
BiaPy is an open source Python library to easily build bioimage analysis pipelines based on deep-learning approaches. The library supports the image processing of 2D, 3D and multichannel microscopy image data. Specifically, BiaPy contains ready-to-use solutions for tasks such as semantic segmentation, instance segmentation, object detection, image denoising, single image super-resolution and image classification, as well as self-supervised learning alternatives.
At present, BiaPy Jupyter notebooks already exporting BioImage.io compatible models are accessible through the BioImage Model Zoo. A future expansion of the current offer by adding a variety of models, including transformers, is expected. The integration of BiaPy in the BioImage Model Zoo aims to enhance the library’s visibility, foster greater collaboration, and serve the community better by increasing the variety of advanced image processing approaches, which significantly empowers the field of BioImage Analysis.


by Beatriz Serrano-Solano
We’re happy to share our latest resource: the AI4Life factsheet! This document provides a comprehensive overview all the important outputs and accomplishments of the project at a glance.
We’ll continuously update this Factsheet to ensure it remains a current resource. Whether you’re preparing a presentation, seeking project insights, or diving into the project’s accomplishments, this Factsheet is what you are looking for.
Feel free to explore and use the AI4Life Factsheet in your outreach activities!


by Beatriz Serrano-Solano
AI4Life recently participated in a series of Machine Learning mini-hackathons hosted by NFDI4DataScience at ZB MED in Cologne (Germany). Our team engaged in two different sessions, aiming to define the Machine Learning lifecycle and to discuss the metadata required for each step.
Machine Learning Lifecycle (21-22 November 2023)
Throughout the two-day event, our objectives revolved around defining the lifecycle steps, creating a graphical representation and fostering compliance with FAIR principles. To extend the discussion to the broader community, the outcomes have been presented at the RDA FAIR4ML Interest Group.
Metadata for Machine Learning (23-24 November 2023)
This session focused on mapping metadata, datasets and applications across various platforms like the DOME registry, Bioimage.io, OpenML, and schema.org, significantly contributing to standardizing ML metadata.
AI4Life’s participation in these mini-hackathons underlines the project’s commitment to enhancing the BioImage Model Zoo models specification to make them interoperable with resources outside the imaging community.
These events carried out during the Machine Learning hackathon at ZB MED sponsored by NFDI4DataScience. NFDI4DataScience is a consortium funded by the German Research Foundation (DFG), project number 460234259.


by Caterina Fuster-Barceló
Introducing the BioImage.IO Chatbot, a game-changer for the bioimage analysis community. This cutting-edge AI-driven assistant is revolutionizing how biologists, bioimage analysts, and developers interact with advanced tools. The BioImage.IO Chatbot excels in delivering personalized responses, code generation, and execution, as demonstrated by various usage examples.
The BioImage.IO Chatbot draws from diverse sources, including databases like ELIXIR bio.tools, documentation from different tools and softwares such as deepImageJ or ImJoy, and the BioImage Model Zoo documentation, ensuring tailored, context-aware answers. The result? Personalized responses that cater to users’ unique requirements.
Distinguished by its versatility, the chatbot adeptly handles both simple and technical queries, ensuring that it remains a valuable asset to users of all backgrounds. What’s more, we are enthusiastic about fostering a community-driven ecosystem. We encourage individuals to integrate their documentation and data sources into our knowledge base, thereby enriching the experience for everyone.
As we prepare for the upcoming beta testing phase (sign up here), join us in witnessing how the BioImage.IO Chatbot is reshaping the landscape of bioimage analysis. Be a part of this transformative journey!
For a detailed overview, check out our preprint.


by Beatriz Serrano-Solano
After a highly successful 2nd General Assembly in Heidelberg, AI4Life took advantage of the gathered members and organized a Hackathon & Solvathon. The event was held from October 10th to 13th, 2023, with a total of 25 participants.
Two parallel events ran during this period:
Both tracks achieved remarkable outcomes. Stay tuned for the next event!

Pictures by Ayoub El Ghadraoui

A new video has been released through our YouTube channel. We asked the participants of the recent Hackathon & Solvathon for a word that describes AI4Life. Hear what they said below!

by Beatriz Serrano-Solano
The AI4Life project held its 2nd General Assembly on October 9th and 10th, 2023, bringing together computational experts and life scientists from various fields to discuss the project’s progress and future goals.
The assembly opened with a welcome from the Euro-BioImaging director, John Eriksson and Euro-BioImaging Bio-Hub director, Antje Keppler.
Anna Kreshuk and Florian Jug, the AI4Life Scientific Coordinators, discussed AI4Life’s objectives, including democratizing AI-based methods, establishing standards for submission, storage, and FAIR (Findable, Accessible, Interoperable, and Reusable) data and models. They also highlighted the need for open calls and empowering common platforms with AI integration.
The keynote speaker was Gergely Sipos, who presented “Computing and AI in EOSC”, discussing opportunities for collaboration. Sipos emphasized the role of EGI, an international e-infrastructure for research and innovation, and its mission to provide computing power, data storage, and training services to the scientific community. He also introduced iMagine, another Horizon Europe-funded project that makes heavy use of EGI resources. Together, AI4Life, iMagine, and EGI decided to explore ways to team up and re-use each others’ technology stacks.
The rest of the meeting was structured from two different angles:
1) Updates from partners: describing the interaction with each other, analysing dependencies within the project, summarising the activities during the first year, and the outlook for the one to come;
2) Updates from Work Packages: describing the goals, deliverables and milestones (submitted and upcoming), and the interactions between work packages. Special focus was given to the question of whether needs have changed since the grant for AI4Life was written. Work Packages also updated the audience with success stories, achievements, goal blockers, pain points, and challenges.
We would like to thank all the participants for their contributions and we are looking forward to an exciting second year of FAIR AI with AI4Life!

by Teresa Zulueta-Coarasa
Artificial Intelligence (AI) methods have revolutionised the analysis of biological images, but their performance depends on the data the models are trained with. Therefore, to develop, benchmark, and reproduce the results of AI methods, developers need access to high-quality annotated data.
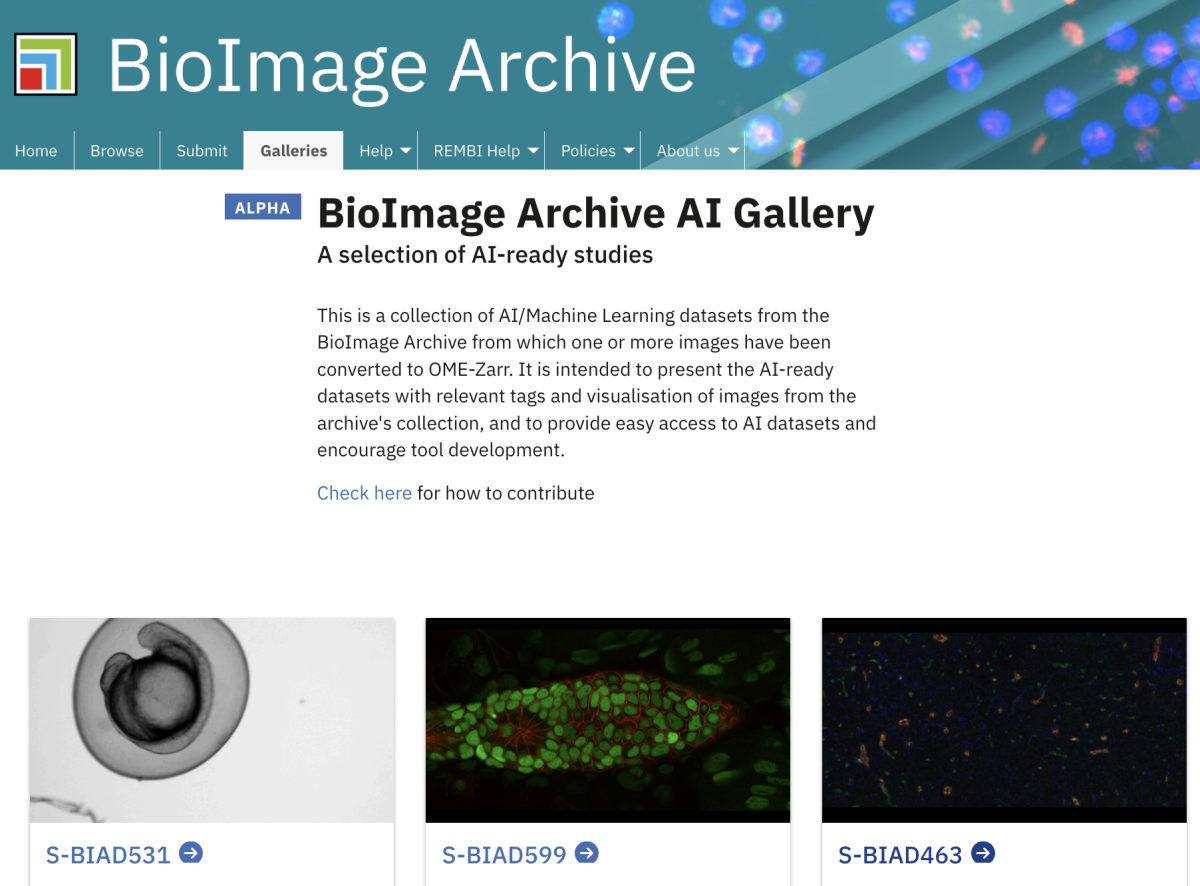
One of the missions of AI4Life is to democratise the access to well-annotated datasets which are standardised to facilitate their reuse, and presented in a manner that is useful to the community. As part of this effort the BioImage Archive has launched a gallery of datasets that can be explored in-browser without the need to download the images and annotations. Each dataset is presented in a consistent way, following community metadata standards that include information such as the biological application of a dataset, what type of annotations a dataset contains, the licence the data are under or what models have been trained using this dataset. Furthermore, because all images are converted from different formats into the cloud-ready file format OME-Zarr, there is potential for analysing these datasets in the cloud.
The BioImage Archive team plans to keep enriching this collection with more datasets over time, with the aim of establishing a community resource that can empower the development of new AI methods for biological image analysis.

by Jeremy Metz, Beatriz Serrano-Solano and Wei Ouyang
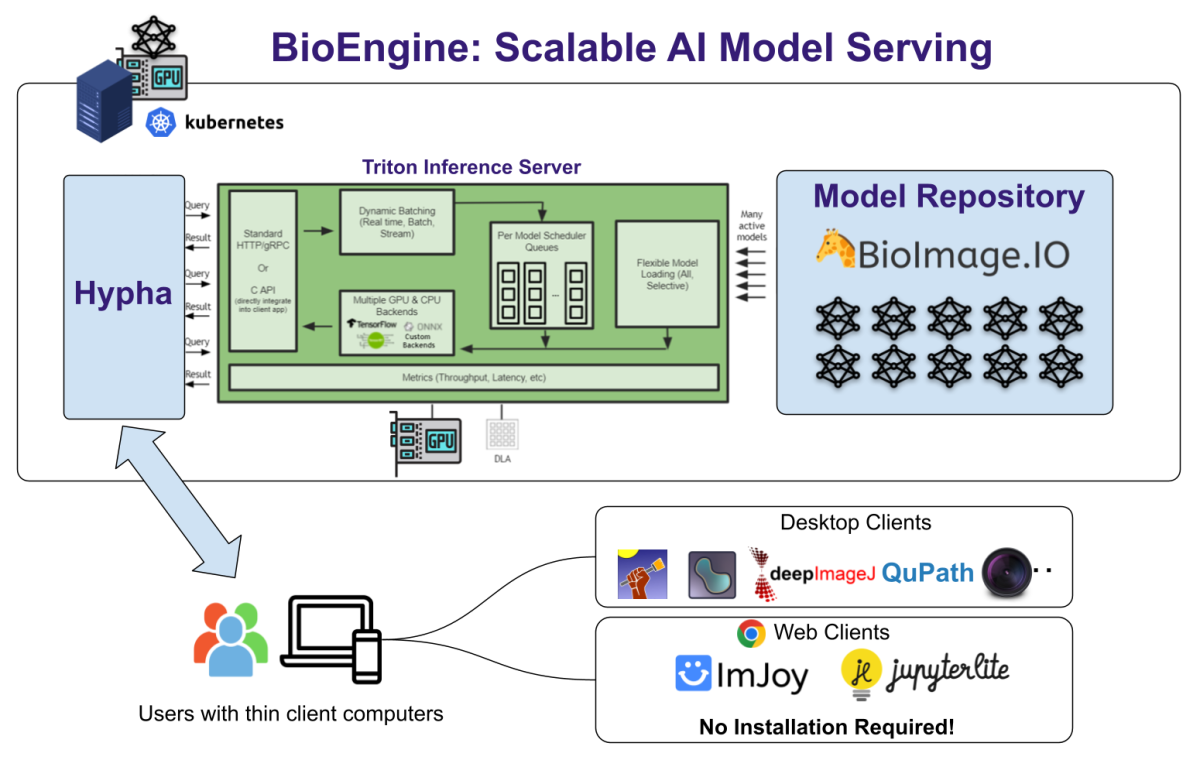
In a major step toward democratizing AI in life sciences, the AI4Life consortium announces the launch of BioEngine—a scalable, cloud-based infrastructure that powers the BioImage Model Zoo. Designed to be accessible to both experts and novices, BioEngine aims to revolutionize the way bioimage analysis is conducted.
The escalating growth of data in life sciences has revealed the limitations of conventional desktop applications used for bioimage analysis. These local solutions are increasingly inadequate to handle high-throughput data and sophisticated applications like AI-driven image analysis. Users often face challenges with large data sets, complex hardware requirements, and intricate software dependencies that make these tools cumbersome to use and difficult to deploy. Additionally, existing machine learning model zoos often necessitate a level of expertise in programming and model selection, making them inaccessible to a wider audience. All these challenges collectively point to the need for a more efficient, scalable, and user-friendly solution for AI model serving.
Enter BioEngine, a state-of-the-art cloud infrastructure designed to simplify the complex landscape of bioimage analysis. BioEngine powers the BioImage Model Zoo, allowing users to test-run pre-trained AI models on their own images without requiring any local installation. Its cloud-based approach means you can easily connect BioEngine to existing software platforms like Fiji, Icy, and napari, thereby eliminating the need to install multiple dependencies.
For the developers in our community, BioEngine serves as a cloud platform designed to cater to many users while judiciously using limited GPU resources. It features a simple API that can be accessed via HTTP or WebSocket, offering a seamless experience for running models in the cloud. This API can be effortlessly integrated into Python scripts, Jupyter notebooks, or web-based applications.
By employing BioEngine, both experts and non-experts can overcome the challenges associated with traditional desktop-based solutions and enter an era of streamlined, accessible and scalable bioimage analysis.
Efficiently serving a variety of models on limited GPU resources required the development of a unique framework that can manage dynamic model execution and scheduling.
BioEngine is the result of a concerted collaboration primarily between KTH Royal Institute of Technology and the BioImage Archive Team at EMBL-EBI, under the auspices of the AI4Life consortium. The platform is built as an extension of the Hypha framework, a robust RPC-based communication hub that orchestrates containerized components for a seamless and efficient operation.
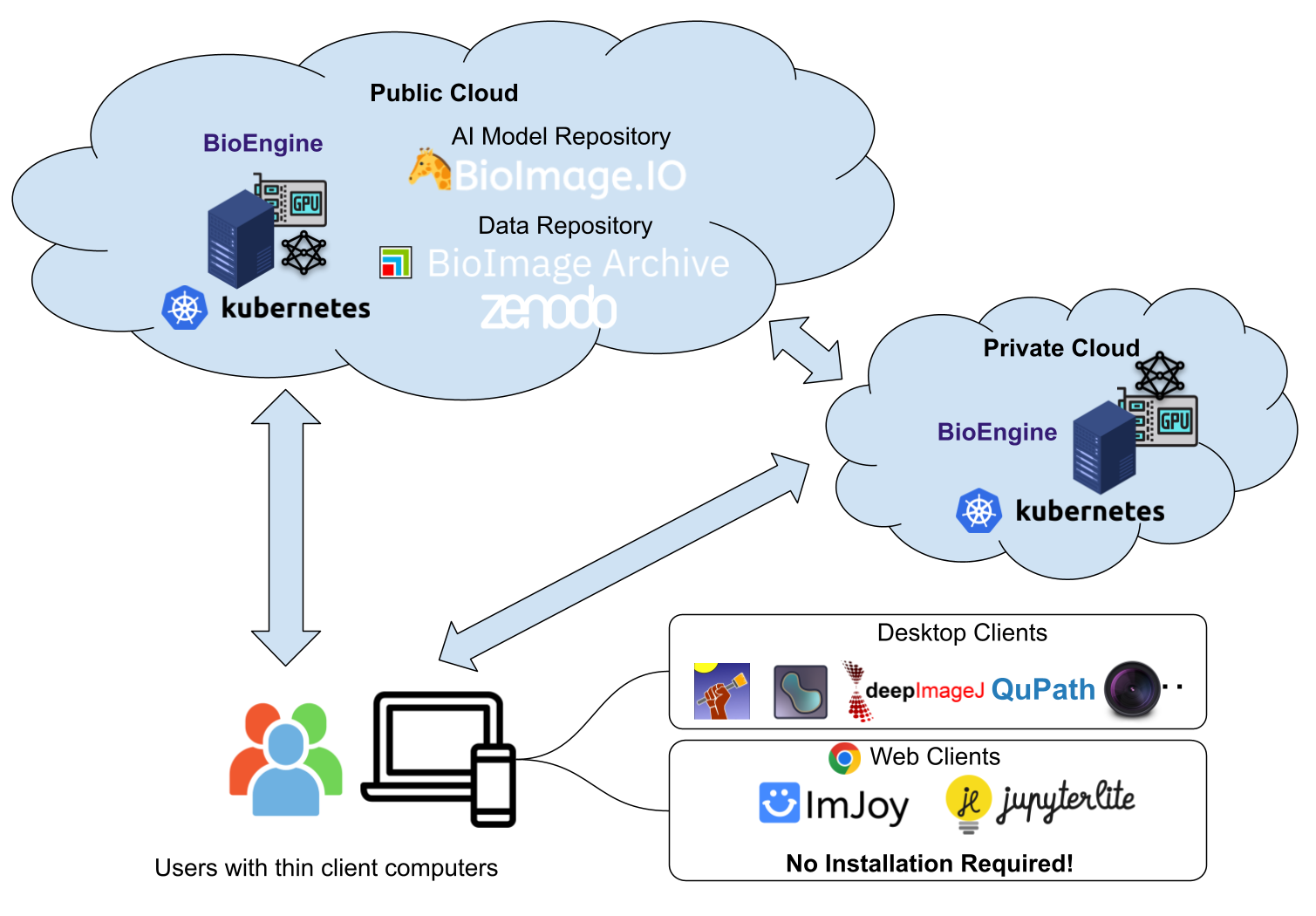
BioEngine provides both HTTP and WebSocket-based RPC APIs. Developers can effortlessly integrate these APIs into web and desktop apps, thereby streamlining the interaction between software and the cloud-based AI models.
We are actively working on broadening our deployment offerings:
With the support the EU Horizon research infrastructure grant, we are committed to promoting accessibility and lowering barriers to advanced bioimage analysis:
We encourage everyone to try BioEngine and provide feedback (via image.sc forum, GitHub issues, or our contact form). Your input is crucial for the continual refinement of this revolutionary platform.
BioEngine offers an easy-to-use API for running models, simplifying software design and deployment.
This API can be integrated into Python scripts, Jupyter notebooks, or web-based applications, giving you the flexibility to adapt BioEngine to your own projects.
We are actively working on an easily deployable package for institutional use and aim to improve stability and scalability.
For more information, please refer to our API documentation.
With BioEngine, we are democratizing advanced bioimage analysis by offering cloud-based, plug-and-play AI solutions that can be effortlessly integrated into existing software ecosystems, thereby accelerating both research and real-world applications in life sciences.