
Author: Pasi Kankaanpää

Introducing the AI4Life Help Desk: Your resource hub for support and information
- Post author By Pasi Kankaanpää
- Post date 31/08/2023

Introducing the AI4Life Help Desk: Your resource hub for support and information
by Caterina Fuster-Barceló
We’re excited to introduce a brand-new addition to the AI4Life project website that will make your experience even more enriching and informative. We’ve just launched the AI4Life Help Desk, a comprehensive resource hub designed to cater to all your queries, support needs, and information requirements.
What is the AI4Life Help Desk?
The AI4Life Help Desk is your go-to destination for accessing a wealth of resources that will enhance your understanding of the AI4Life project and the BioImage Model Zoo. Whether you’re a seasoned user or just getting started, our Help Desk is here to assist you every step of the way.
What’s inside the Help Desk?
- Contact Us: Reach out through Image.sc/bioimageio, GitHub, or our user-friendly Typeform for tailored support.
- Training and Documentation: Access BioImage Model Zoo guides, Galaxy Training Network resources, and AI4Life tutorials.
- FAQs: Get quick answers to common questions about AI4Life and the BioImage Model Zoo.
Discover the AI4Life Help Desk today at https://ai4life.eurobioimaging.eu/help!

New section on the AI4Life Website: Explore our latest outputs!
- Post author By Pasi Kankaanpää
- Post date 14/08/2023

New section on the AI4Life Website: Explore Our Latest Outputs!
We are excited to introduce a fresh addition to the AI4Life website dedicated to showcasing our project’s accomplishments in a more accessible manner.
Milestones and Deliverables
AI4Life is a very prolific project, we will publish 25 deliverables and 16 milestones throughout the project’s duration. This newly launched section serves as a hub where you can dig into the specifics of each deliverable and milestone. An interactive timeline provides insight into the projected completion dates of our deliverables, offering a clear roadmap for our ongoing endeavours.
Highlighting Our Scientific Contributions
In this same section, we’ve also gathered our scientific publications. By centralizing these contributions, our aim is to foster knowledge-sharing and encourage collaboration within the wider scientific community.
Your Window into AI4Life’s Advancements
We invite you to explore this new section of our website to gain insight into the work that has defined AI4Life’s journey thus far. By making our outputs easily accessible, we hope to engage you with our work. And, for those that want to stay closely connected to AI4Life, we offer the option to subscribe to our newsletter.

Models developed in AI4Life and Bioimage.io now made available in AIVIA
- Post author By Pasi Kankaanpää
- Post date 14/07/2023

Models developed in AI4Life and Bioimage.io now made available in AIVIA
AI4Life and Leica are announcing their collaboration to make deep-learning models developed by the bioimage community available to a wider user community through integration into Leica’s AIVIA software.
In a commitment to the scientific community, AI4Life and Leica Microsystems join forces to help researchers to leverage AI in complex experiments. AI4Life, coordinated by Euro-BioImaging, is a Horizon Europe-funded project that brings together the computational and life science communities. Its goal is to empower life science researchers to harness the full potential of Artificial Intelligence (AI) methods for bioimage analysis – and in particular microscopy image analysis, by providing services, and developing standards aimed at both developers and users.
One of the consortium’s objectives is to build an open, accessible, community-driven repository (the BioImage Model Zoo) of FAIR pre-trained AI models and develop services to deliver these models to life scientists. Together with their community partners, the consortium ensures that models and tools are interoperable with Fiji, ImageJ, Ilastik and other open-source software tools.
In the meantime, Leica Microsystems has cultivated a valuable connection with the AI4Life project and the BioImage Model Zoo. Most recently, the Leica team had the opportunity to meet the people behind the AI4Life project at a workshop organized by the Euro-BioImaging Industry Board (EBIB). It was there, through their participation, that they realized the power of their combined resources.
Widely recognized for optical precision and innovative technology, Leica Microsystems supports the imaging needs of the scientific community with AIVIA, their advanced Al-powered image analysis software. AIVIA is a complete 2-to-5D image visualization and analysis platform designed to allow researchers to unlock insights previously out of reach. Through the joined effort, BioImage Model Zoo models can now also be easily integrated into Leica’s commercial software AIVIA.
“We see this initiative as an opportunity to tie in AIVIA with the scientific community and to make work done by the community accessible in a convenient way to AIVIA users.” said Constantin Kappel, Manager AI Microscopy and Insights at Leica Microsystems.
To achieve this, Leica is taking the models that are published in BioImage.io and converting them to their own AIVIA Model repository format for interoperability with AIVIA. From a small number of models, the offer will gradually be increased.
This further supports the aim of AI4Life to lower the barriers for using pre-trained models in image data analysis for users without substantial computational expertise or those with existing licences, who might rely on commercial solutions.
This is a great example of how resources curated by academia and available in open access can be of interest to the imaging industry. The source of the models are recognized directly in AIVIA, a nice testimony to industry-academia collaboration and a nice endorsement of the models curated by the BioImage Model Zoo.
“We much appreciate the interest of Leica. Models relevant to AIVIA will be directly available to users, which is a testimony to fruitful industry-academia collaboration and a great endorsement of the utility of the BioImage Model Zoo. We hope many other software tools will follow Leica’s lead and also start benefitting from this community resource we are currently building” said Florian Jug, one of the scientific coordinators of AI4Life.
“Bioimage.io is supported by AI4Life. AI4Life has received funding from the European Union’s Horizon Europe research and innovation programme under grant agreement number 101057970.”
More information:
https://www.aivia-software.com/post/pancreatic-phase-contrast-cell-segmentation-bioimage-io
https://www.aivia-software.com/post/hpa-cell-segmentation-bioimage-io
https://www.aivia-software.com/post/hpa-nucleus-segmentation-bioimage-io
https://www.aivia-software.com/post/b-subtilist-bacteria-segmentation-bioimage-io

First AI4Life Open Call: Announcement of selected projects
- Post author By Pasi Kankaanpää
- Post date 04/07/2023

First AI4Life Open Call:
Announcement of selected projects
by Florian Jug & Beatriz Serrano-Solano
The first AI4Life Open Call received an impressive response, with a total of seventy-two applications. It proved to be an incredible opportunity for both life scientists seeking image analysis support and computational scientists eager to explore the evolving landscape of AI methodologies. In this blog post, we announce the awarded projects and invite you to join us behind the scenes as we explore the selection process that determined which projects have been selected.
Awarded projects
First things first, here is the list of titles of the selected projects (in alphabetical order):
- Analysis of the fiber profile of skeletal muscle.
- Atlas of Symbiotic partnerships in plankton revealed by 3D electron microscopy.
- Automated and integrated cilia profiling.
- Identifying senescent cells through fluorescent microscopy.
- Image-guided gating strategy for image-enabled cell sorting of phytoplankton.
- Leaf tracker plant species poof.
- SGEF, a RhoG-specific GEF, regulates lumen formation and collective cell migration in 3D epithelial cysts.
- Treat CKD.
The projects are diverse, covering scientific topics ranging from Plant Biology, Physiology, Metabolism, Cell Biology, Molecular Biology, Marine Biology, Flow Cytometry, Medical Biology, Regenerative Biology, Neuroscience, etc. The researchers who have proposed the projects come from the following countries: France (2x), Germany, Italy, Netherlands, Portugal, and the USA (2x).
How did the review procedure work?
1. Eligibility checks
The selection procedure started with internal eligibility checks. Is the project submitted completely? Is the information complete and telling a complete story that is fit for external reviews? At this stage, we only had to drop 10 projects of a grant total of 72 submitted projects. Our intention was to only filter projects that drew an incomplete picture and leave the judgement of the scientific aspects to our reviewers.
2. Reviewing procedure
After assembling a panel of 16 international reviewers (see list below), we distributed anonymized projects among them. All personal and institutional information was removed, only leaving project-relevant data to be reviewed. We aimed at receiving 3 independent reviews per project, requiring each review to review about 11 projects total.
Here is the list of questions we asked our reviewers via an electronic form:
- Please rank the following statements from 1 (Likely not) to 5 (Likely):
- The proposed project is amenable to Deep Learning methods/approaches/tools.
- Does the project have well-defined goals (and are those goals the correct ones)?
- A complete solution to the proposed project will require additional classical routines to be developed.
- The project, once completed, will be useful for a broader scientific user base.
- The project will likely require the generation of significant amounts of training data.
- This project likely boils down to finding and using the right (existing) tool.
- Approaches/scripts/models developed to solve this project will likely be reusable for other, similar projects.
- The project, once completed, will be interesting to computational researchers (e.g. within a public challenge).
- The applicant(s) might have a problematic attitude about sharing their data.
- Data looks as if the proposed project might be feasible (results good enough to make users happy).
- Do you expect that we can (within reasonable effort) improve on the existing analysis pipeline?
- What are the key sub-tasks the project needs us to improve?
- What would you expect will it take (in person-days again) to generate sufficient training data?
- Do suitable tools for this exist? What would you use?
- Once sufficient training data exists, what would you expect is the workload for AI4Life to come up with a reasonable solution for the proposed project? Please answer first in words and then (further below) with the minimum and maximum number of days you expect this project to take.
- What is your estimated minimum number of days for successfully working on this project?
- What is your estimated maximum number of days for successfully working on this project?
- On a scale from 1 to 10, how enthusiastic are you about this project?
Due to the unforeseen unavailability of some reviewers, we ended up with about 2.7 reviews per project, with some projects receiving 2 but most projects receiving all 3 desired reviews.
3. Scoring projects according to reviewer verdicts
We first aggregated all reviews per project by averaging numerical values and concatenating textual evaluations. We then developed three project scores: a quality score (main metric), a total effort score, and a slightly more subjective excitingness score.
- The quality score was computed by taking a weighted average of the evaluations we received. I.e., questions (1-a) to (1-k) from above. (Note: not for all questions higher values are better. We have of course first inverted the “low-is-better” ones to make all values compatible.)
- The effort score was taking the (minimum) time estimates for label data generation and successfully completing the project, and computing a value corresponding to the estimated total person-month to completion.
- The excitingness score simply is the average of the values received as answers to question 8.
The final score was computed by: 0.75*(quality/effort) + 0.25*excitingness
This formula favors projects that are estimated to be conducted in less time, which is in line with our aim to help more individuals through the AI4Life Open Calls.
4. Final decisions by the Open Call Selection Committee
- After anonymized scoring of all projects, we have added the applicants’ identities and institutions back into the final decision matrix.
- We have prepared ourselves to break ties and potentially remove better-ranked projects for the sake of having a higher diversity. To our surprise, the top-ranked projects showed a wonderful diversity, making this step unnecessary.
- The final decision was taken by the Open Call Selection Committee. With the members of the committee (see below), we have re-lived all steps of the Open Call process, from the application, and reviewing, to the final grading stage. After some stability analysis (i.e., after changing the weights for the weighted sums in the procedure outlines above and noticing that the best projects remained rather stably top-ranked), the Committee decided to simply select as many of the best-evaluated projects as we could fit into the AI4Life time budget for this round of Open Calls. This led to a total number of 8 selected projects.
- Seeing the extraordinary quality of many of the submitted projects, it was clear to us that many more than 8 projects would deserve to receive support. We have therefore decided to put a sizeable number of additional projects on a waiting list, hoping that we can engage more helping hands.
Who was involved in the review process?
- Marvin Albert
- Ashesh Ashesh
- Damian Dalle Nogare
- Joran Deschamps
- Vera Galinova
- Kate Gill
- Iván Hidalgo
- Varun Kapoor
- Dominik Kutra
- Thibault Lagache
- Arrate Muñoz Barrutia
- Anil Kumar Mysore Badarinarayana
- Constantin Pape
- Anirban Ray
- Pablo Rodríguez Pérez
- Daniel Sage
- Ignacio Arganda-Carreras
- Jan Funke
- Florian Jug
- Anna Kreshuk
- Emma Lundberg
- Aastha Mathur
- Damian Dalle Nogare
- Florian Jug
And now? What’s next?
The selected projects will be assigned to our AI4Life experts waiting to support them. All other projects are offered a space in the AI4Life Bartering Corner, a new section soon to appear on our website, where projects will be showcased to computational experts who can reach out to the proposing parties and engage in a fruitful collaboration.
If you did not apply to the first Open Call, we invite you to do so at the beginning of 2024. Subscribe to our newsletter, we will inform you when the next call opens.
Additionally, if you are interested to put any open analysis problem you have on our Bartering Corner, please fill out this form.
If you need help quicker, we recommend Euro-BioImaging’s Web Portal, where you can access a network of experts in the field of image analysis. Please note that this service may involve associated costs, but access funds for certain research topics are available through initiatives such as ISIDORe.

Outcomes of the hackathon on web and cloud infrastructure for AI-powered bioimage analysis
- Post author By Pasi Kankaanpää
- Post date 22/06/2023

Outcomes of the Hackathon on Web and Cloud infrastructure for AI-powered bioimage analysis
by Caterina Fuster-Barceló
The AI4Life Hackathon on Web and Cloud Infrastructure for AI-Powered BioImage Analysis recently took place at SciLifeLab in Stockholm, Sweden. Organized by Wei Ouyang of KTH Sweden, in partnership with AI4Life and Global BioImaging, the event aimed to bring together experts in the field to discuss and design advanced web/cloud infrastructure for bioimage analysis using AI tools. Participants from both academia and industry worldwide attended, showcasing platforms like BioImage Model Zoo, Fiji, ITK, Apeer, Knime, ImJoy, Piximi, Icy, and deepImageJ. Read more in this article written by the project partners in FocalPlane.

AI4Life teams up with the Galaxy Training Network to enhance training resources
- Post author By Pasi Kankaanpää
- Post date 20/06/2023
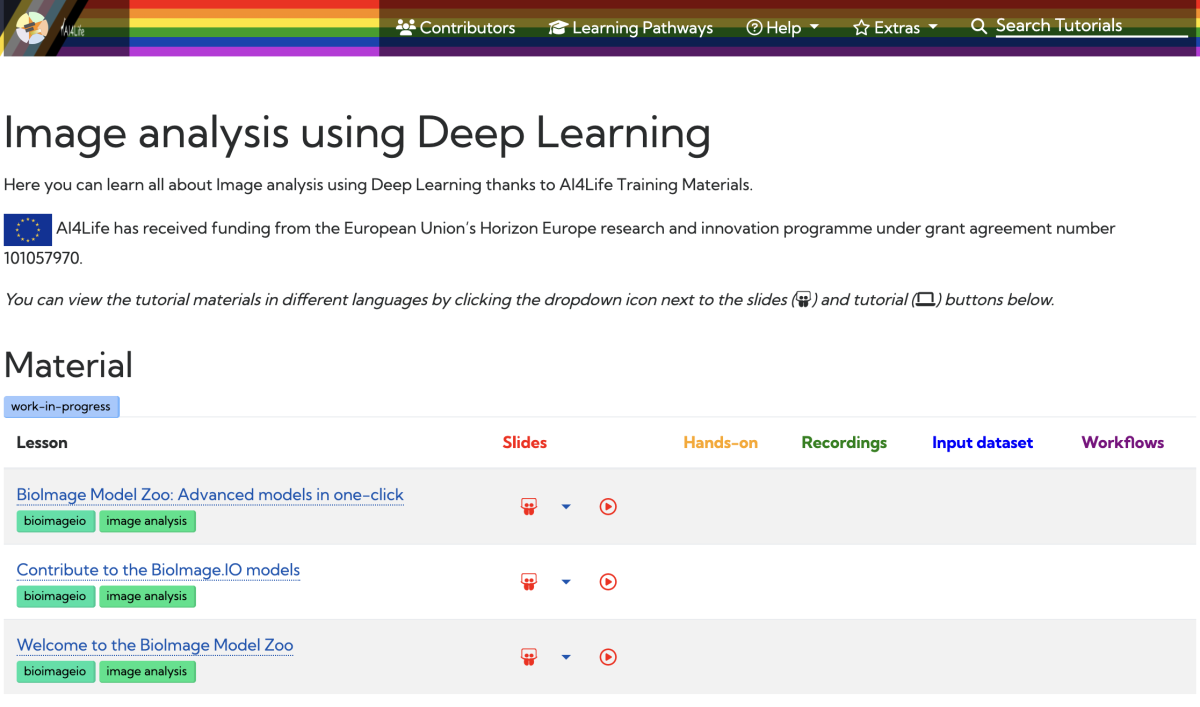
AI4Life teams up with Galaxy Training Network (GTN) to enhance training resources
by Caterina Fuster-Barceló
In an exciting collaboration, AI4Life has joined forces with the Galaxy Training Network (GTN) project to revolutionize the way researchers access training materials. The GTN, known for its dedication to promoting FAIR (Findable, Accessible, Interoperable, and Reusable) and Open Science practices globally, now incorporates AI4Life to expand its training offerings.
Through this collaboration, BioImage Model Zoo (BMZ) and AI4Life trainers have developed videos and slides to introduce the community to the BMZ, demonstrate proper utilization, and guide contributions. This exciting development allows the BMZ to reach a wider audience within the research community and offers a simplified, visual approach to understanding and utilizing the BMZ.
This collaboration between the BMZ and GTN opens up new opportunities for researchers to access training materials and gain a better understanding of the BMZ’s capabilities. By making the process more accessible and intuitive, the BMZ aims to facilitate its adoption among researchers from diverse backgrounds.
The integration of the BMZ into the GTN project represents a significant advancement in training resources, empowering researchers worldwide and fostering collaboration within the scientific community. Stay tuned for upcoming training materials that will unlock the full potential of the BMZ for your research pursuits.
AI4Life at the 5th NEUBIAS Conference
- Post author By Pasi Kankaanpää
- Post date 26/05/2023
AI4Life at the 5th NEUBIAS Conference
by Estibaliz Gómez-de-Mariscal
The 5th NEUBIAS Conference took place in Porto during the week of May 8th, 2023. It brought together experts in BioImage Analysis for the Defragmentation Training School and the Open Symposium. AI4Life actively participated in the event, contributing to both parts and covering topics from zero code Deep Learning tools, the Bioimage Model Zoo, BiaPy, Segment Anything for Microscopy, among others. Estibaliz Gómez-de-Mariscal has written a post in FocalPlane summarising the discussions and outcomes.

BioImage Model Zoo joins Image.sc forum as a Community Partner
- Post author By Pasi Kankaanpää
- Post date 12/05/2023

BioImage Model Zoo joins Image.sc forum as a Community Partner
by Caterina Fuster-Barceló
The BioImage Model Zoo (BMZ) has been incorporated as a Community Partner of the Image.sc forum, a discussion forum for scientific image software sponsored by the Center for Open Bioimage Analysis (COBA). The BMZ is a repository of pre-trained deep learning models for biological image analysis, and its integration into the Image.sc forum will provide a platform for the community to discuss and share knowledge on a wide range of topics related to image analysis.
The Image.sc forum aims to foster independent learning while embracing the diversity of the scientific imaging community. It provides a space for users to access a wide breadth of experts on various software related to image analysis, encourages open science and reproducible research, and facilitates discussions about elements of the software. All content on the forum is organized in non-hierarchical topics using tags, such as the “bioimageio” tag, making it easy for people interested in specific areas to find relevant discussions.
As a Community Partner, the BMZ joins other popular software tools such as CellProfiler, Fiji, ZeroCostDL4Mic, StarDist, ImJoy, and Cellpose, among others. The partnership means that the BMZ will use the Image.sc forum as a primary recommended discussion channel, and will appear in the top navigation bar with its logo and link.
The Image.sc forum has been cited in scientific publications, and users may reference it using the following citation:
Rueden, C.T., Ackerman, J., Arena, E.T., Eglinger, J., Cimini, B.A., Goodman, A., Carpenter, A.E. and Eliceiri, K.W. “Scientific Community Image Forum: A discussion forum for scientific image software.” PLoS biology 17, no. 6 (2019): e3000340. doi:10.1371/journal.pbio.3000340
The integration of the BMZ into the Image.sc forum will undoubtedly facilitate knowledge-sharing and collaborative efforts in the field of biological image analysis, benefiting researchers, developers, and users alike.
Outcomes of the First AI4Life Open Call
- Post author By Pasi Kankaanpää
- Post date 18/04/2023

Outcomes of the First AI4Life Open Call
by Beatriz Serrano-Solano & Florian Jug
We are thrilled to announce that we received an impressive number of 72 applications to the first AI4Life Open Call!
The first AI4Life Open Call was launched in mid-February and closed on March 31st, 2023. It is part of the first of a series of three that will be launched over the course of the AI4Life project.
AI4Life involves partners with different areas of expertise, and it covers a range of topics, including marine biology, plant phenotyping, compound screening, and structural biology. Since the goal of AI4Life is to bridge the gap between life science and computational methods, we are delighted to see so much interest from different scientific fields seeking support to tackle scientific analysis problems with Deep Learning methods.
We noticed that the most prominent scientific field that applied was cell biology, but we also received applications from neuroscience, developmental biology, plant ecology, agronomy, and marine biology, as well as aspects of the medical and biomedical fields, such as cardiovascular and oncology.
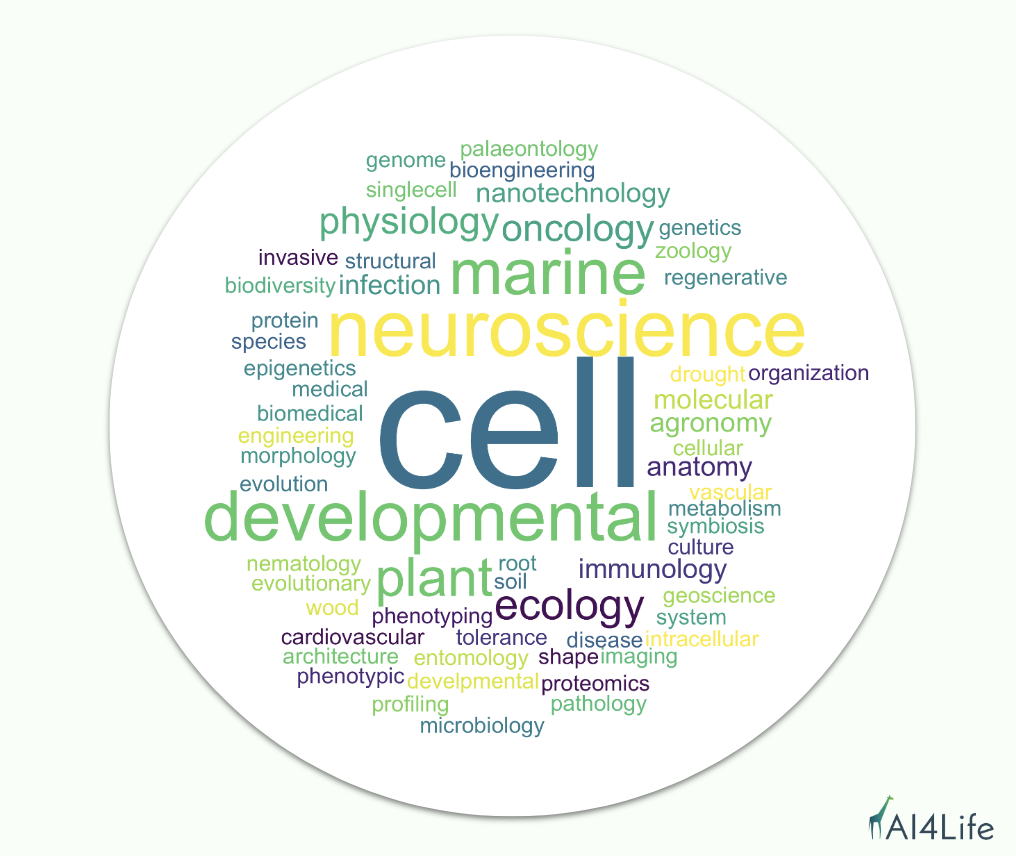
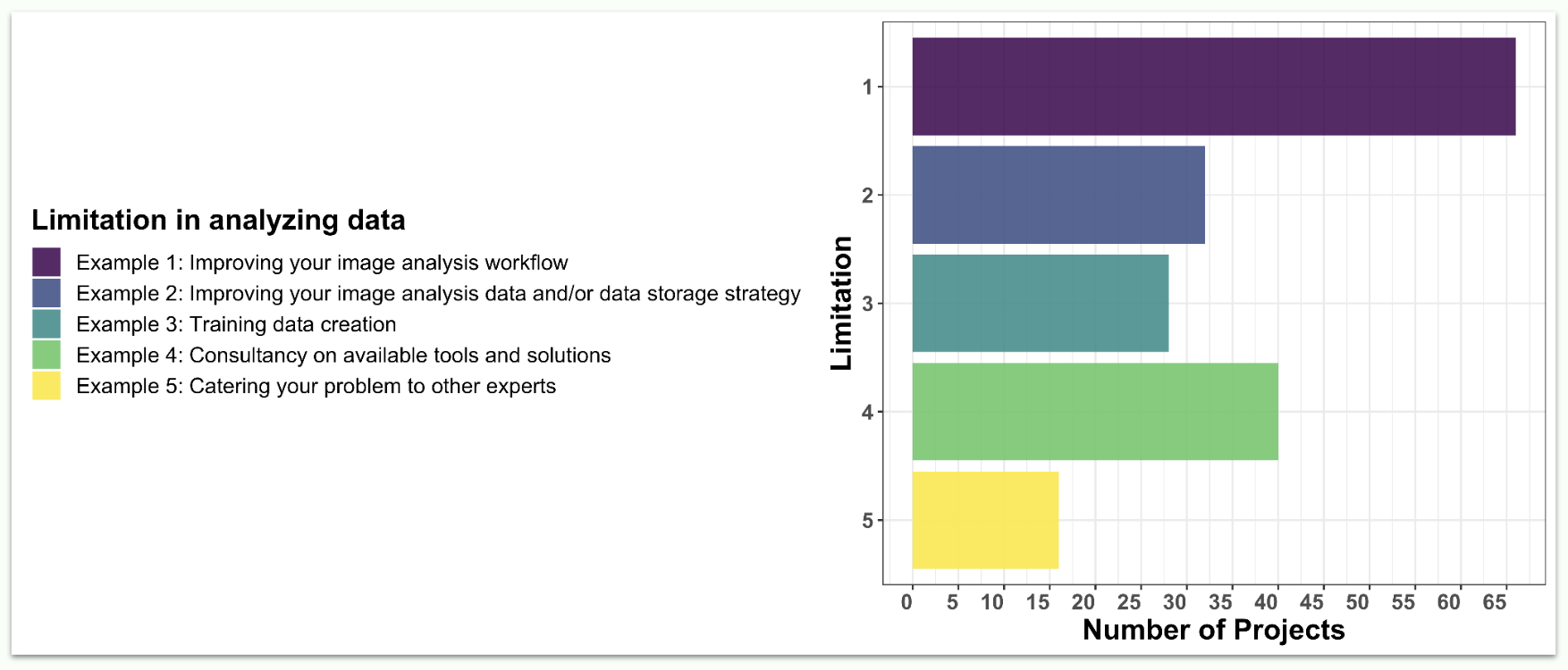
Why did scientists apply? What challenges are they facing?
Applications were classified based on the type of problem that will need to be addressed. We found that improving the applicant’s image analysis workflow was the most common request (67 applications out of 72). This was followed by improving image analysis data and/or data storage strategy, training data creation, and consultancy on available tools and solutions. Of course, we also welcomed more specific challenges that were not falling within any of those categories.
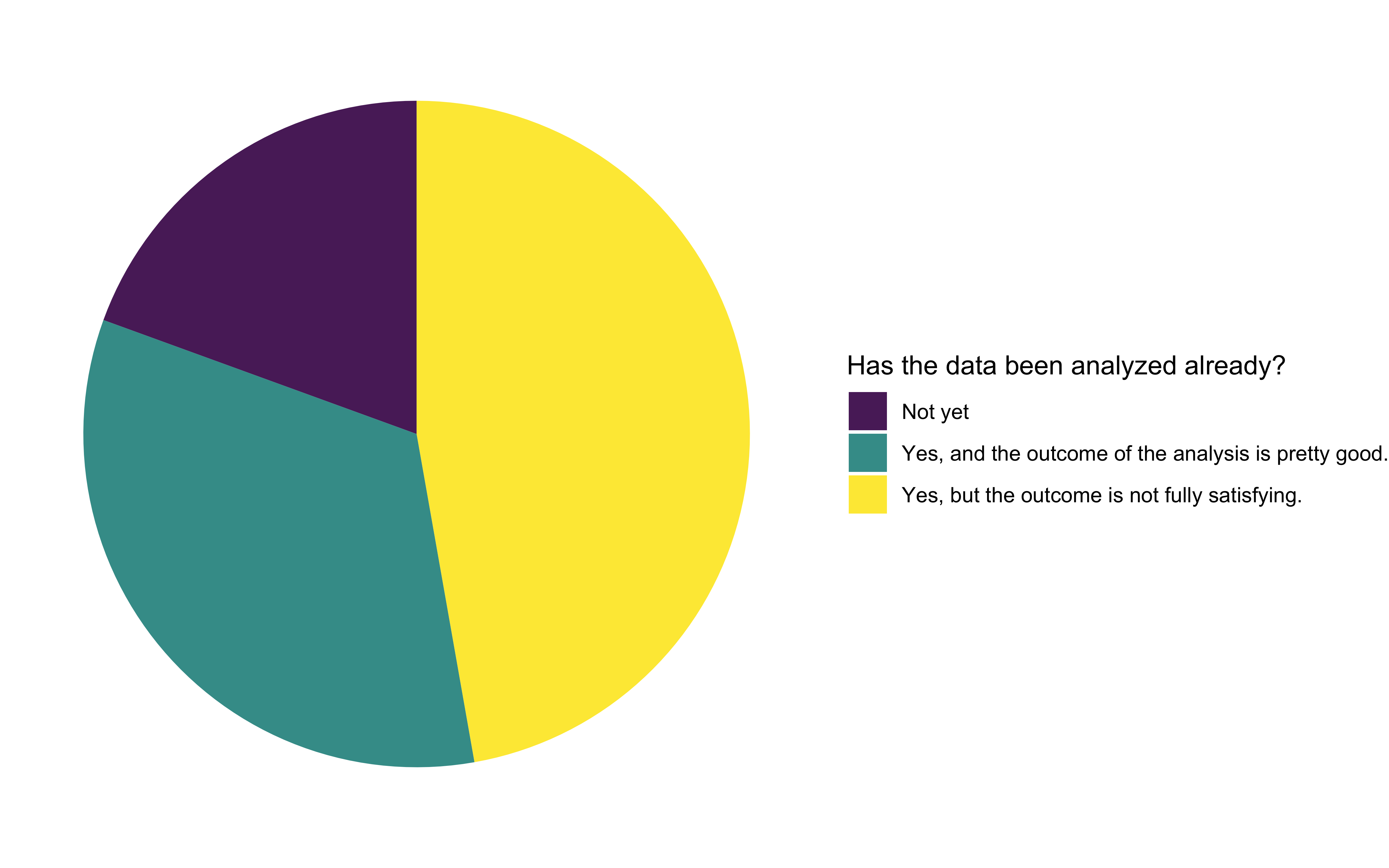
How have applicants addressed their analysis problem so far?
We were pleased to see that most of the applicants had already analyzed the data, but half of them were not fully satisfied with the outcomes. We interpreted this as an opportunity to improve existing workflows. The other half of the applicants were satisfied with their analysis results, but longed for better automation of their workflow, so it becomes less cumbersome and time-consuming. Around 20% of all applications haven’t yet started analyzing their data.
When asked about the tools applicants used to analyze their image data, Fiji and ImageJ were the most frequently used ones. Custom code in Python and Matlab is also popular. Other frequently used tools included Napari, Amira, Qupath, CellProfiler, ilastik, Imaris, Cellpose, and Zen.

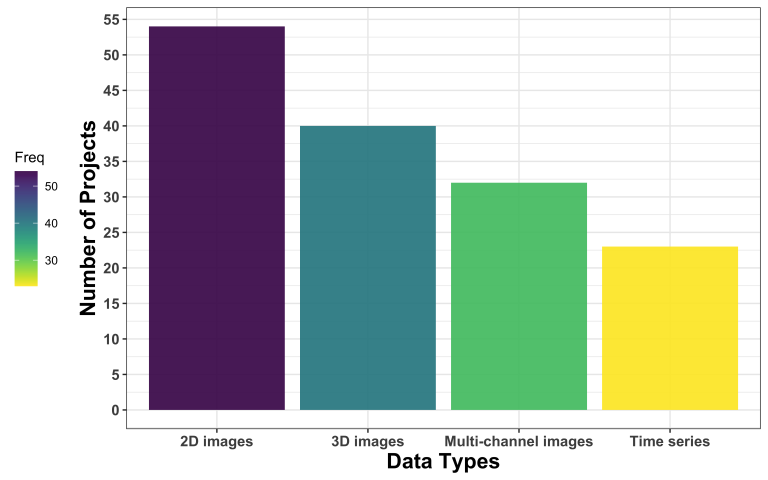
What kind of data and what format do applicants deal with?
The most common kind of image data are 2D images, followed by 3D images, multi-channel images, and time series.
Regarding data formats, TIFF was the most popular one, followed by JPG, which is borderline alarming due to the lossy nature of this format. AVI was the third most common format users seem to be dealing with. CSV was, interestingly, the most common non-image data format.
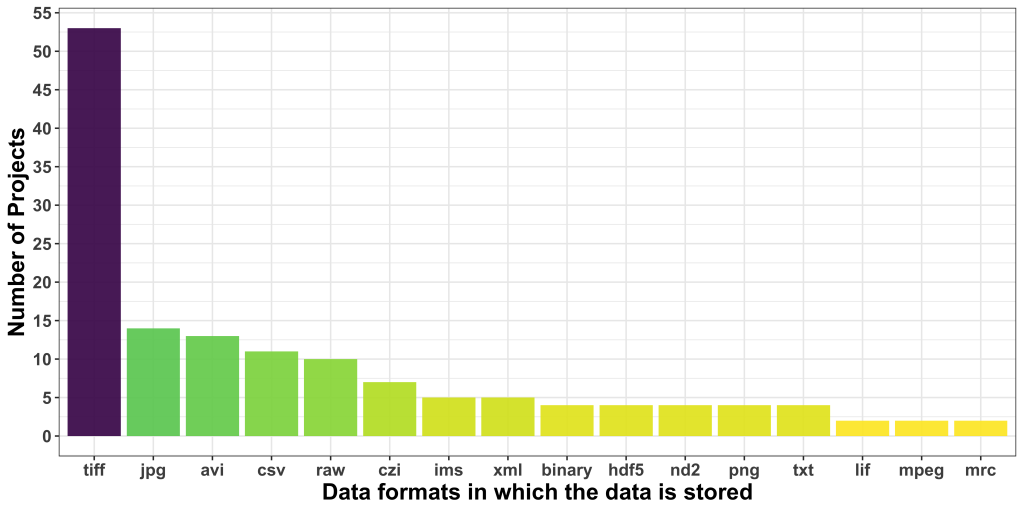
Additionally, we asked about the relevance of the metadata to address the proposed project. 17 applicants didn’t reply, and 24 others did, but do not think metadata is quite relevant to the problem at hand. While this is likely true for the problem at hand, these responses show that the reusability and FAIRness of acquired and analyzed image data is not yet part of the default mindset of applicants to our Open Call.
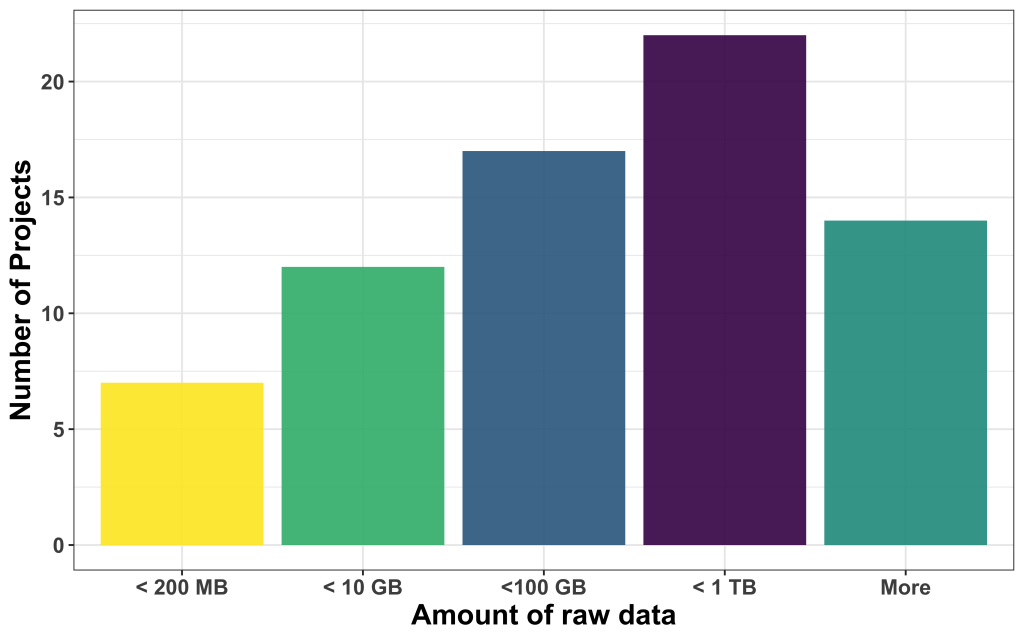
How much project-relevant data do the applicants bring?
We found that most of the data available was large, with most of them in the range between 100 and 1000 GB, followed by projects that come with less than 10 GB, between 10 and 100 GB, and less than 200 MB. 14 projects had more data than 1 TB to offer.
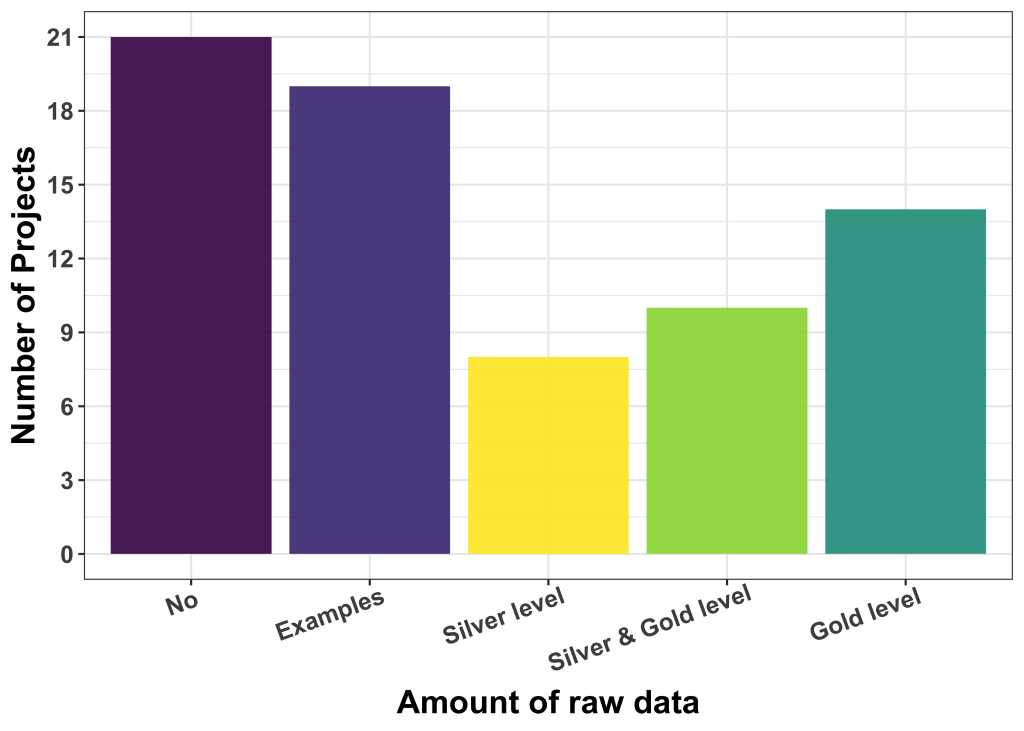
Is ground truth readily available?
We also asked about the availability of labelled data and provided some guidelines regarding the kind and quality of such labels. We distinguished: (i) Silver ground truth, i.e. results/labels good enough to be used for publication (but maybe fully or partly machine-generated, and (ii) gold ground truth, i.e. human curated labels of high fidelity and quality.
The majority of applicants (40) had no labelled data or only very few examples. The rest have silver level (8), a mix of silver and gold levels (10) and gold level (14) ground truth available. The de-facto quality of available label data is, at this point in time, not easy to be assessed, but our experience is that users who believe to have gold-level label data are not always right.
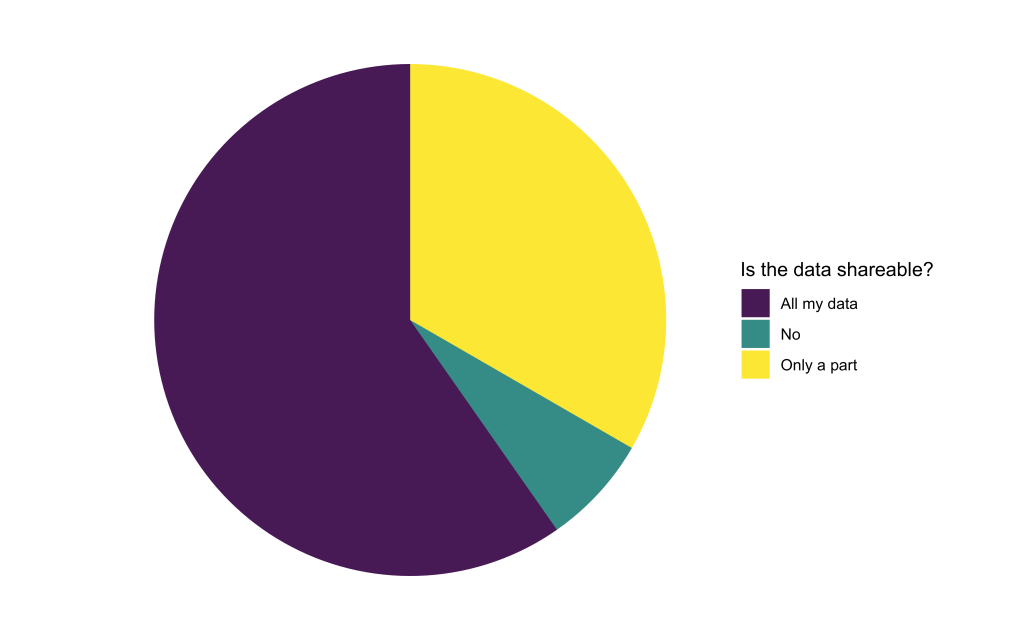
Is the available data openly shareable?
To train Deep Learning models, the computational experts will need access to the available image data. We found that only a small portion of applicants were not able to share their data at all. The rest is willing to share either all or at least some part of their data. When only parts of the data are sharable, reasons were often related to data privacy issues or concerns about sharing unpublished data.
What’s next? How will we proceed?
We are currently undergoing an eligibility check and the pool of reviewers will start looking at the projects in more detail. In particular, they will rank the projects based on the following criteria:
- The proposed project is amenable to Deep Learning methods/approaches/tools.
- Does the project have well-defined goals (and are those goals the correct ones)?
- A complete solution to the proposed project will require additional classical routines to be developed.
- The project, once completed, will be useful for a broader scientific user base.
- The project will likely require the generation of significant amounts of training data.
- This project likely boils down to finding and using the right (existing) tool.
- Approaches/scripts/models developed to solve this project will likely be reusable for other, similar projects.
- The project, once completed, will be interesting to computational researchers (e.g. within a public challenge).
- The applicant(s) might have a problematic attitude about sharing their data.
- Data looks as if the proposed project might be feasible (results good enough to make users happy).
- Do you expect that we can (within reasonable effort) improve on the existing analysis pipeline?
The reviewers will also identify the parts of the project that can be improved, evaluate if deep learning can be of help and provide an estimation of the time needed to support the project.
We will keep you posted about all developments. Thanks for reading thus far! 🙂


