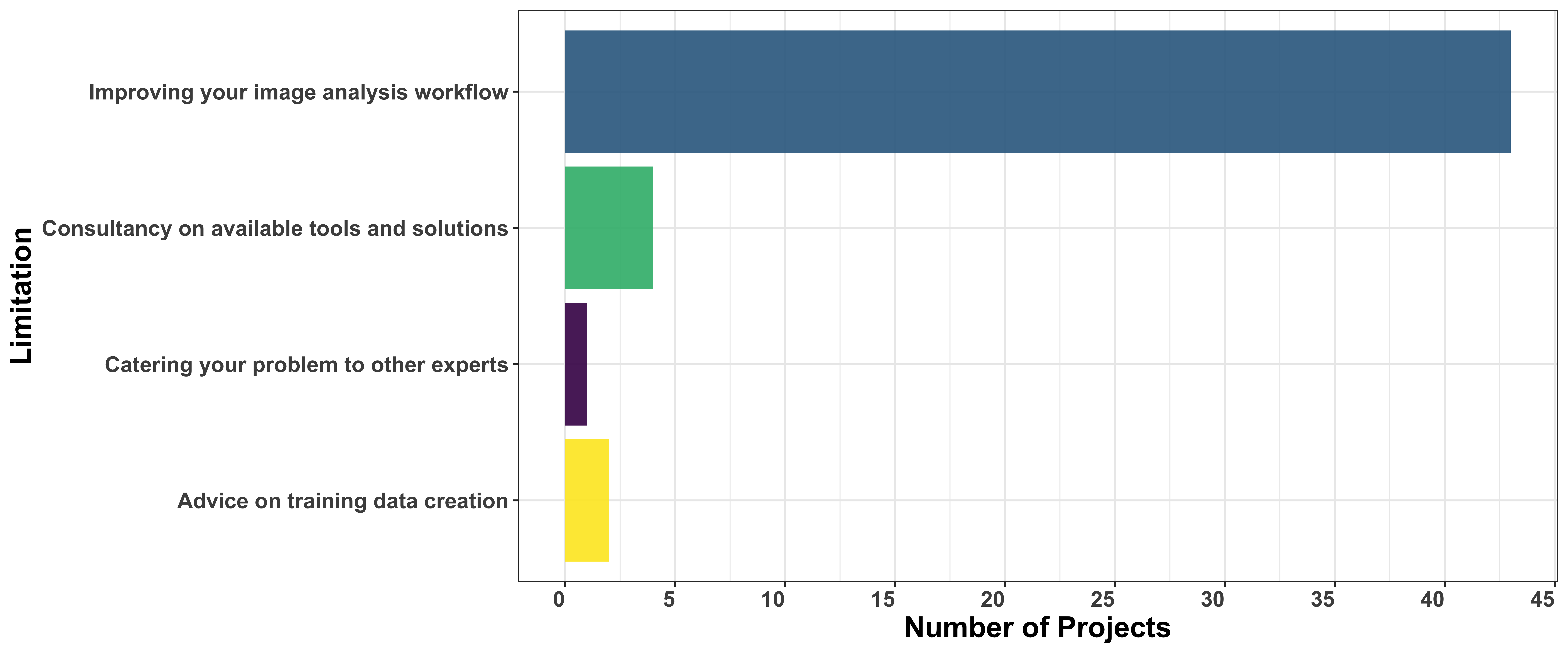
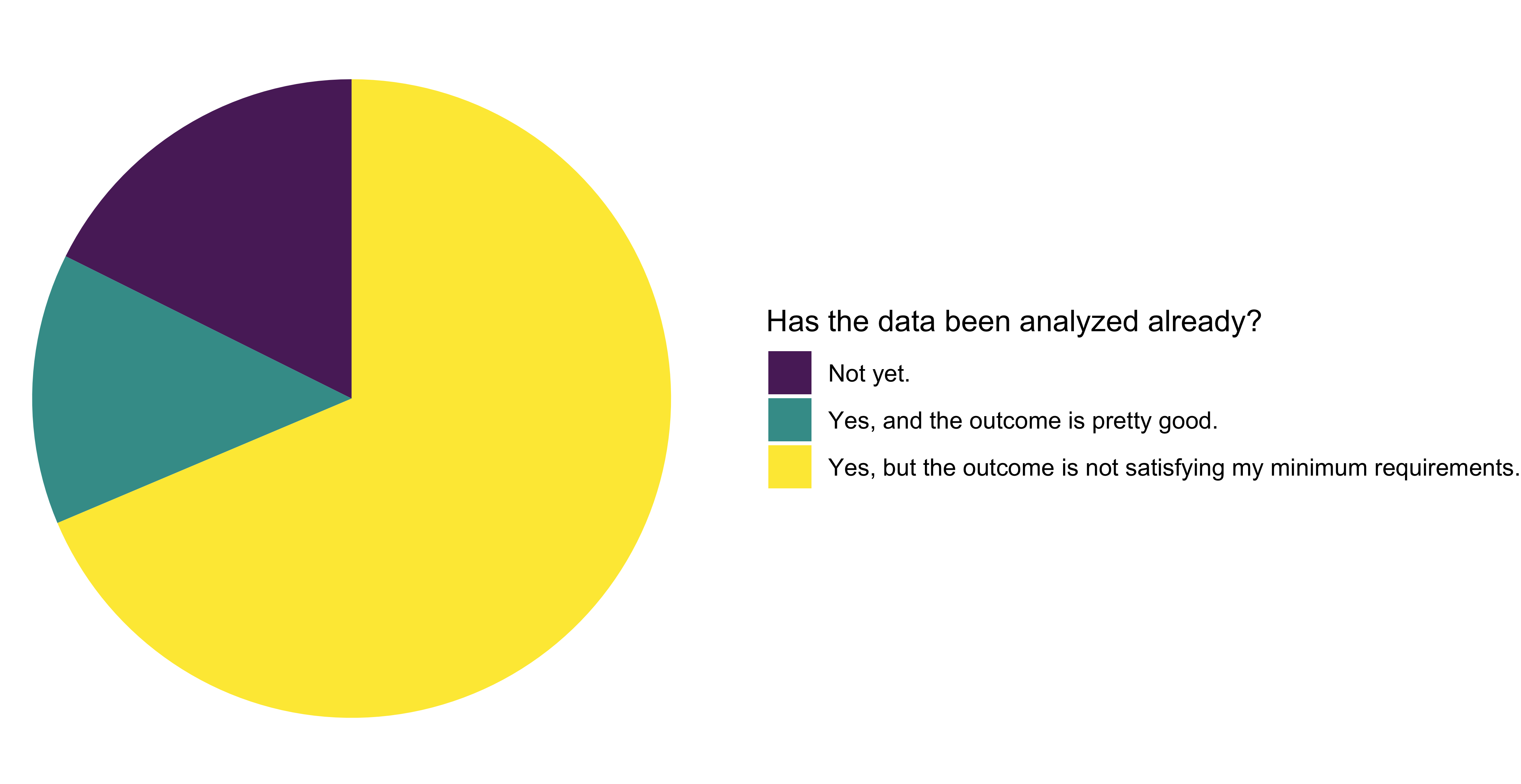
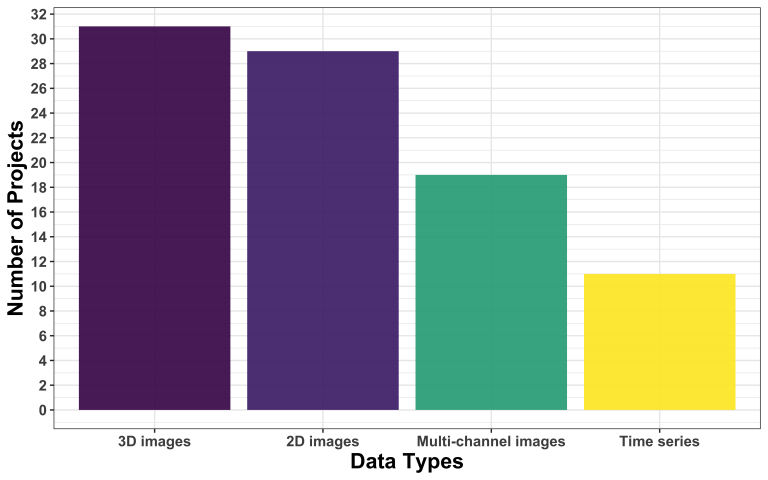
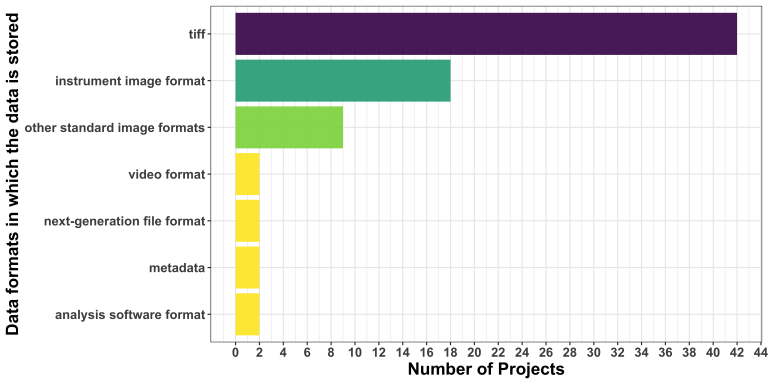
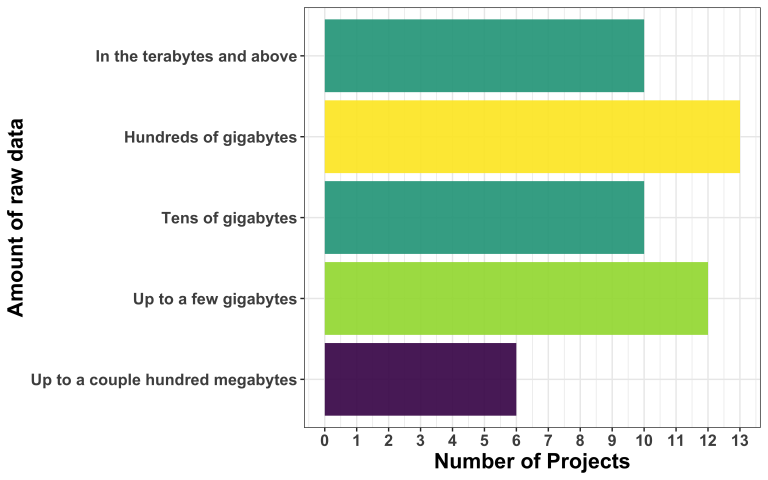
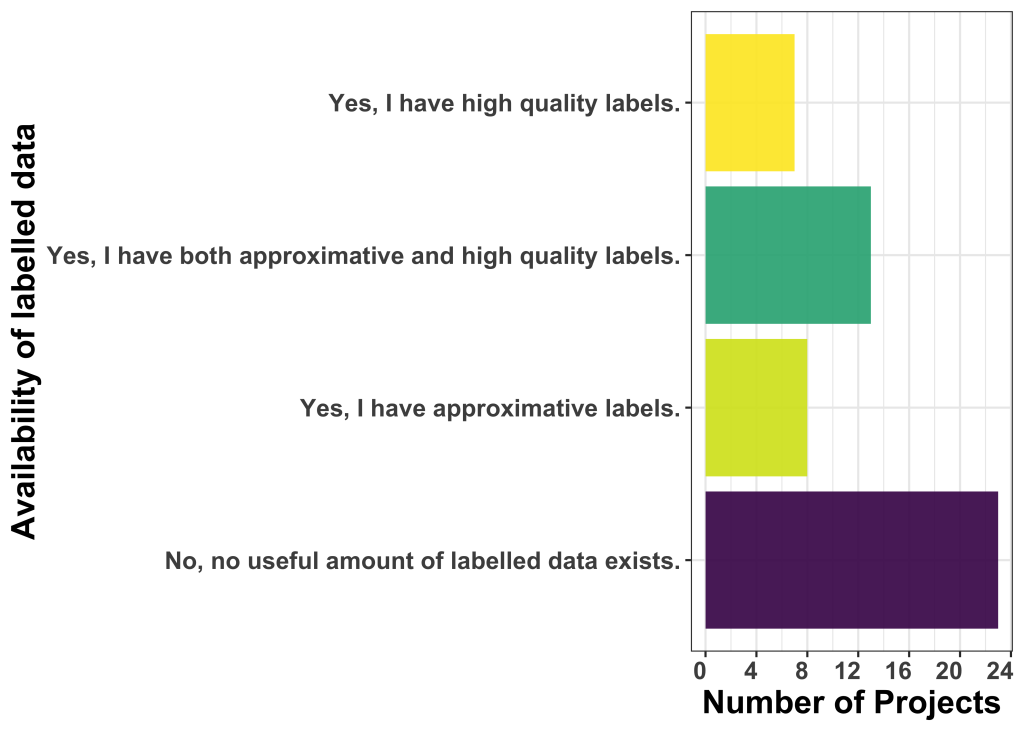
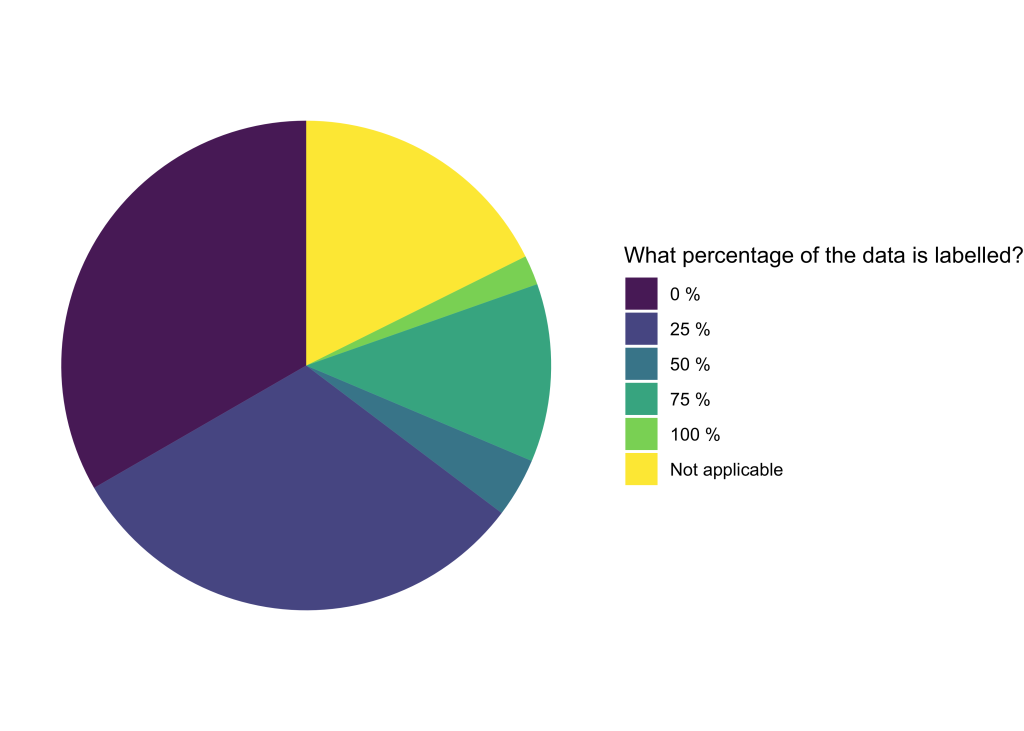
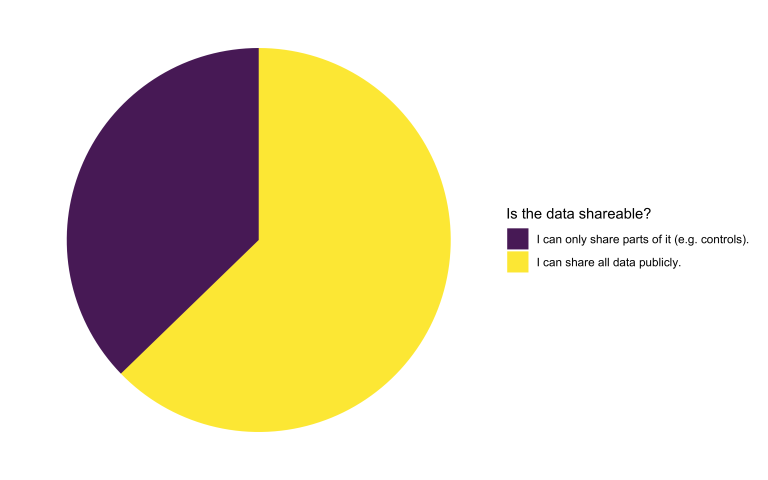
Model uploader
One of the key focus areas was to put the final touches to a new model uploader, aimed at simplifying the process of uploading models to the BioImage Model Zoo. The uploader will no longer rely on external platforms like Zenodo; instead, models will be hosted internally and authentication will be requested for contributors who want to upload a model. One of the teams worked to simplify authentication procedures and optimize model uploads to S3 by integrating Google authentication, providing a unified system that would enhance the overall user experience.
Infrastructure improvement
Teams dedicated their efforts to refining Continuous Integration (CI) processes, which have now been migrated to the collection-bioimage-io GitHub repository. The uploader now triggers the CI workflow, automating the process of pushing models to the designated storage location on S3.
JupyterHub and DL4MicEverywhere
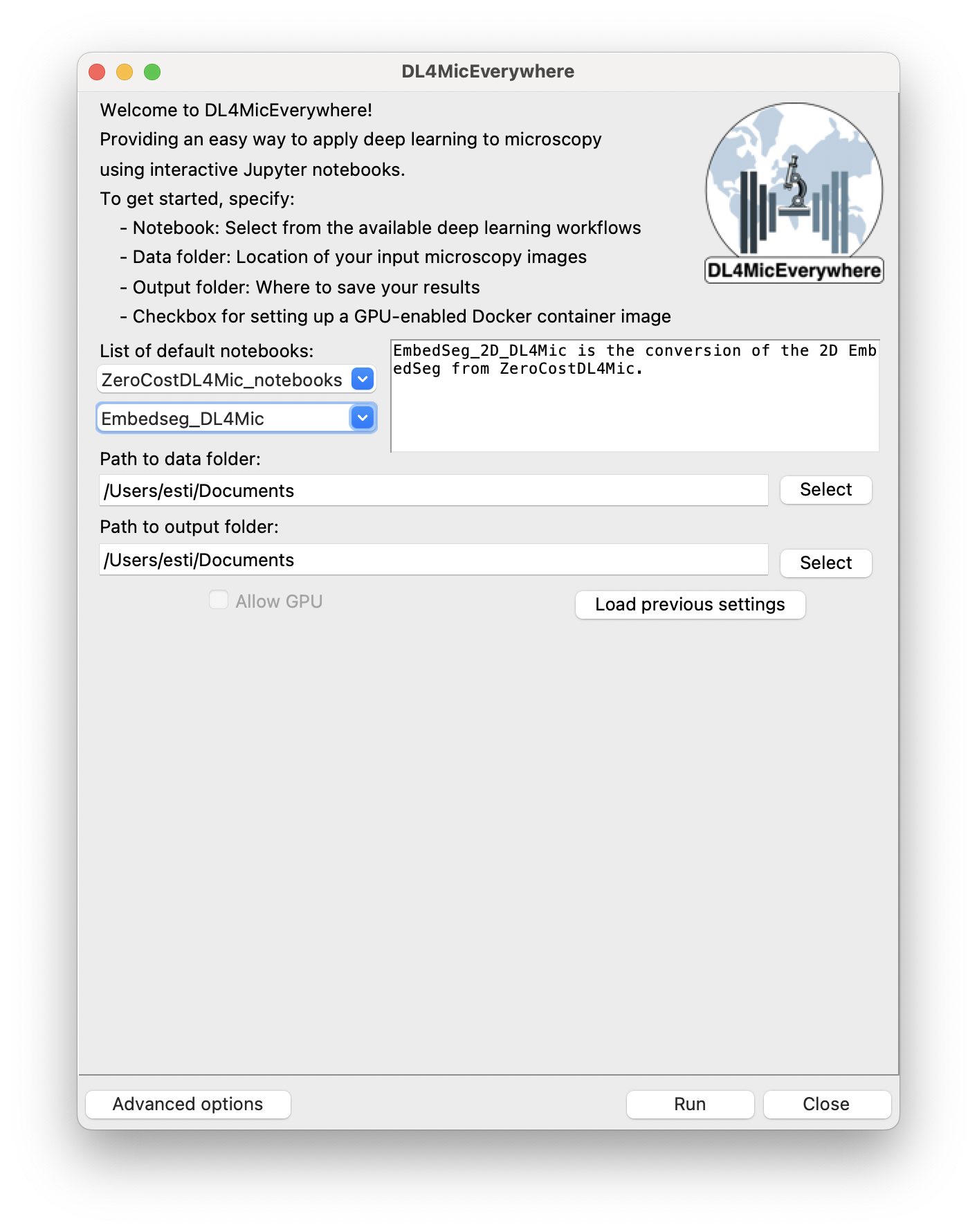
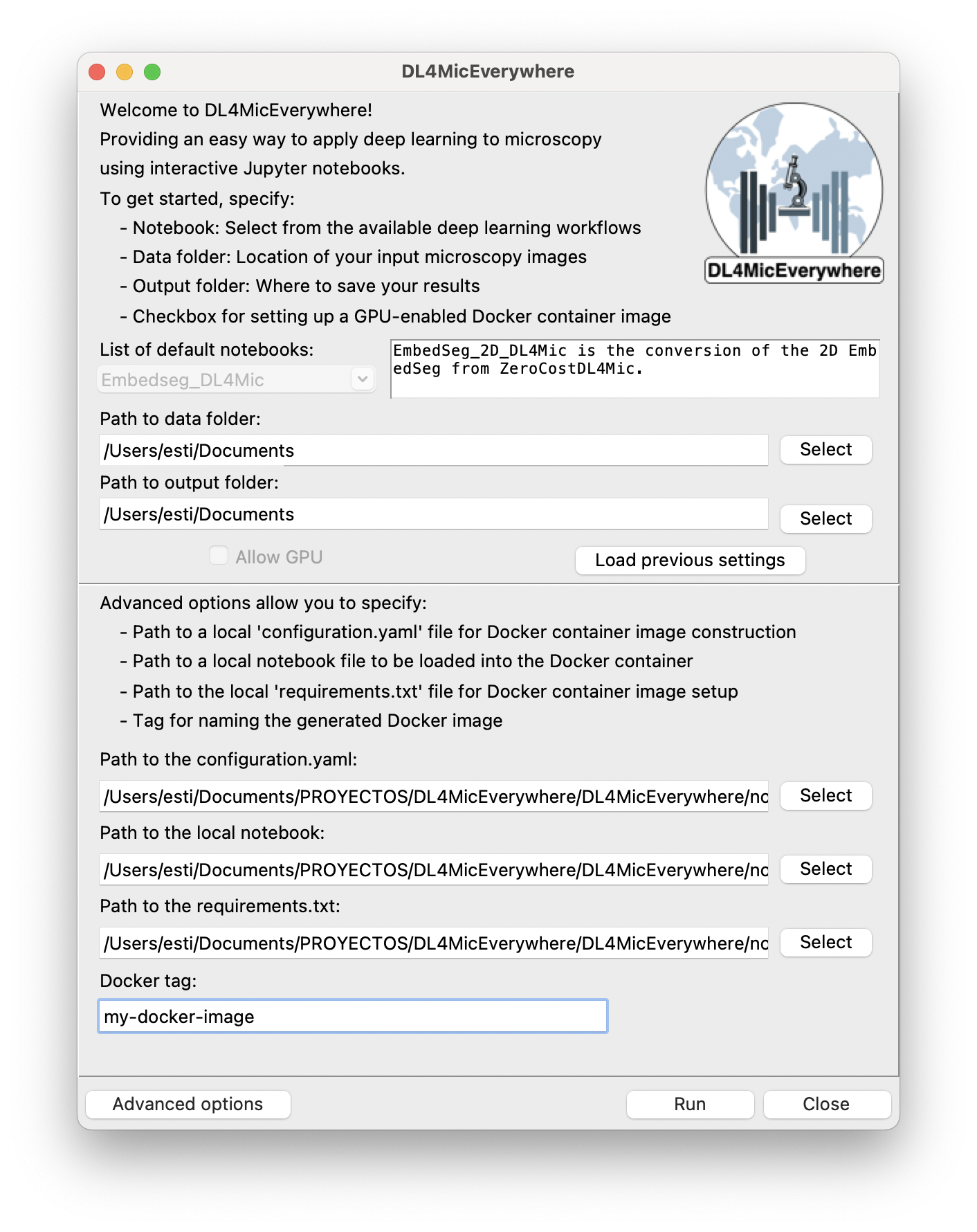
Another focus area involved transitioning the infrastructure for JupyterHub from Google Colab to Google Cloud, providing users with a more robust and flexible environment.
Model quantization
Model quantization allows making networks smaller and faster without loss of precision. We held discussions describing current state of the art.. As an example of the performance gains, a 3D Unet model from the BioImage Model Zoo reduced the inference of a batch of images from 60 ms to 30 ms.
Hypha launcher
Hypha can now launch BioEngine (triton-server) on a Slurm cluster using Apptainer. Additionally, a service-id option is now implemented in the BioEngine web client to easily switch the execution backend to high-performance computing (HPC) environments. Furthermore, BioEngine can now be launched on desktop environments.
Model export to new specifications
This team focused on exporting models using the new specifications. Additionally, the team explored approaches to export CellPose models.
Documentation enhancement
This project was split into two phases: firstly, restructuring the current documentation and secondly, creating new needed documentation for the BioImage Model Zoo. Community input and feedback are highly encouraged in this project!
Second Open Call
The deadline for the second Open Call was March 8th. During the hackathon, we had the opportunity to engage with all the reviewers, many of whom were participating in the event. Projects were assigned to each reviewer, kickstarting officially the review process.
Thank you to everyone who contributed either onsite or online. It was a pleasure to work with this engaged group of people. And thank you to the AI4Health innovation cluster for supporting this event. We look forward to meeting you at the next event!